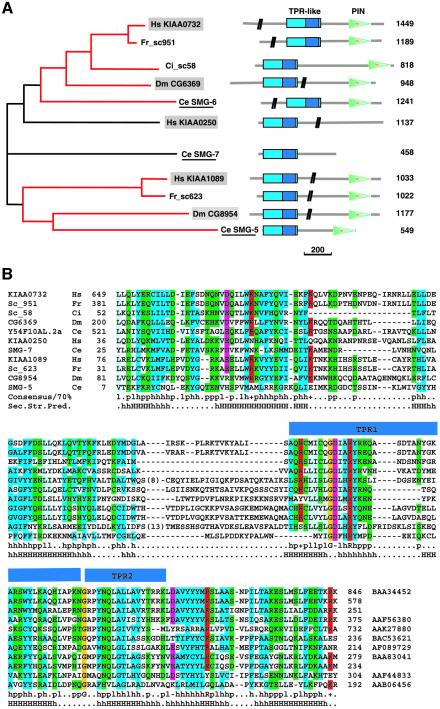
Fig. 3. Phylogenetic tree and domain organization of the SMG5–7 proteins. (A) The two identified orthologous groups are shown in red. Only one and a half TPR repeats can be identified automatically and are indicated in dark blue within the conserved helical N-terminal domain (cyan). The C.elegans sequences are underlined and are known to be involved in NMD (Cali et al., 1999; Page et al., 1999; Anders et al., 2003). Human and Drosophila sequences characterized in this study are shown in gray boxes. The vertical black bars on the schematic of the protein domains indicate that part of the protein sequence is not drawn to scale. The number of amino acid residues is shown on the right. Scale bar: 200 amino acids. TPR, tetratricopeptide repeats; PIN, PilT-N-terminus; Ce, Caenorhabditis elegans; Ci, Ciona intestinalis; Dm, Drosophila melanogaster; Fr, Fugu rubripes; Hs, Homo sapiens. (B) Multiple sequence alignment of the TPR-like region. The accession number of each sequence is indicated, as well as the species and the start and end points of the sequences. For the sequences of C.intestinalis and F.rubripes, no accession numbers are shown, as the predictions were performed directly on the genomic sequences using GeneWise (Birney et al., 1996). The consensus in 70% of the sequences is shown below the multialignment; h, l, p, +, – represent hydrophobic, aliphatic, polar, positive and negative residues, respectively. Aliphatic residues are highlighted in cyan, hydrophobic in blue, polar in green, negative in pink, and positive in red. The conserved glycines are highlighted in orange. The secondary structure prediction (Sec.Str.Pred.) is taken from the consensus of the multiple alignment (H, helices predicted with an accuracy >82%; h, helices predicted with an accuracy ≤82%]. The blue bars above the sequences show the position of the one and a half TPR repeats identified by Clissold and Ponting (2000).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.