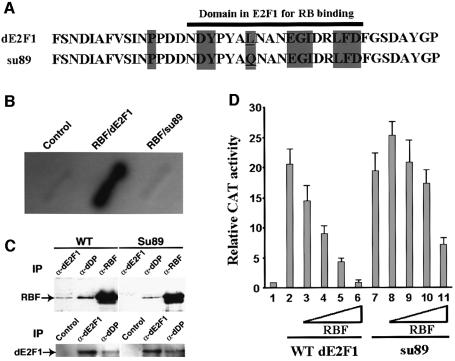
Fig. 1. Molecular characterization of the dE2F1su89 mutation. (A) The point mutation (underlined) in the dE2F1su89 C-terminus is shown. Amino acids that are conserved between Drosophila and mammalian E2Fs are shaded in gray, and the Rb-binding domain of dE2F1 is indicated with a black bar. (B) dE2F1su89 did not interact with RBF in a yeast two-hybrid interaction assay. β-Gal activity on patches of yeast transformed with control plasmids or plasmids encoding RBF, wild-type dE2F1 or dE2F1su89 (su89) is shown as indicated. (C) Endogenous dE2F1su89 protein did not form a complex with RBF. Extracts from wild-type (WT) or dE2F1su89 (su89) embryos were immunoprecipitated with anti-dE2F1, dDP and RBF antibodies as indicated, followed by western blot with an antibody against RBF or dE2F1 as indicated to determine the association between the endogenous RBF and dE2F1su89 proteins. (D) dE2F1su89 can activate transcription normally but is defective in its regulation by RBF. SL2 cells were transfected with no dE2F1 (1), a constant amount of wild-type dE2F1 (2–6) or dE2F1su89 (7–11), and increasing amounts of RBF as indicated. The averages of normalized CAT activities from two independent experiments are shown.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.