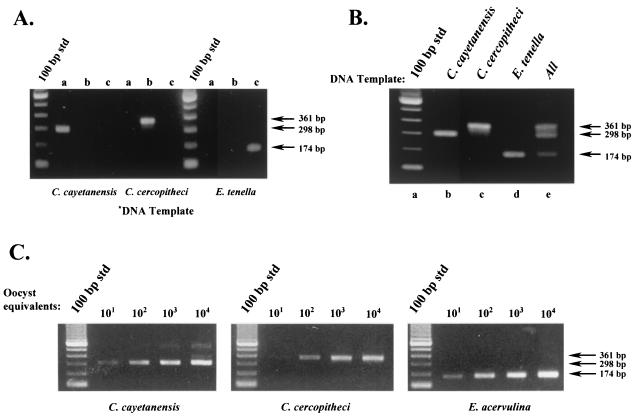
FIG. 2.
Differential detection of Cyclospora and Eimeria species by using SNP primers. (A) Evaluation of SNP primer specificity. Primary PCR amplicons from C. cayetanensis, C. cercopitheci, and E. tenella were generated with the F1E-R2B primer pair. Portions (1 μl) of these products were subjected to nested-amplification reactions with their respective SNP primers as follows: lane a, SNP primer CC719; lane b, SNP primer PDCL661; and lane c, SNP primer ESSP841. (B) Multiplex PCR amplification with SNP primers to differentially detect Cyclospora and Eimeria species. Portions (1 μl) of the primary amplicons from C. cayetanensis, C. cercopitheci, and E. tenella were used as a template in a multiplex amplification reaction containing all of the SNP primers listed in Table 2 as follows: lane b, C. cayetanensis; lane c, C. cercopitheci; lane d, E. tenella; and lane e, all templates. Lane a is a standard 100-bp DNA ladder. (C) Detection sensitivity of the SNP multiplex PCR protocol. Primary PCR amplicons were prepared with the primer pair F1E-R2B. DNA templates were prepared from C. cayetanensis, C. colobi, and E. acervulina by using either intact oocysts spotted onto FTA filters (C. cayetanensis and E. acervulina) or the equivalent amount of lysate (C. colobi). Portions (3 μl) of the primary amplicons were used as a template in a multiplex amplification reaction containing all of the SNP primers as described in panel B.