Abstract
The filamentous soil bacterium Streptomyces coelicolor undergoes a complex cycle of morphological differentiation involving the formation of an aerial mycelium and the production of pigmented antibiotics. We have developed a procedure for generating insertional mutants of S. coelicolor based on in vitro transposition of a plasmid library of cloned S. coelicolor DNAs. The insertionally mutated library was introduced into S. coelicolor, and transposon insertions were recovered at widely scattered locations around the chromosome. Many of the insertions revealed previously uncharacterized genes, and several caused novel mutant phenotypes, such as altered pigment production, enhanced antibiotic sensitivity, delayed or impaired formation of aerial hyphae, and a block in spore formation. The sporulation mutant harbored an insertion in one of three adjacent genes that are apparently unique to Streptomyces but are each represented by at least 20 paralogs at dispersed locations in the chromosome. Individual members of the three families often are found grouped together in a characteristic arrangement, suggesting that they have a common function.
Streptomyces are filamentous soil bacteria that undergo a complex cycle of morphological differentiation (1–3). The cycle begins with a spore, which germinates to give rise to a branching network of multinucleoid hyphae known as the substrate mycelium. Morphological differentiation commences with the formation of specialized aerial hyphae, which grow into the air away from the surface of the colony. Later these aerial hyphae undergo septation into uninucleoid compartments and metamorphose into chains of gray-pigmented spores. The onset of differentiation frequently is associated with the production of pigments, antibiotics, and other secondary metabolites (3, 4).
Relatively few morphological mutants have been described in Streptomyces, and even fewer have been characterized at the molecular level. One reason for slow progress is that cloning genes identified by chemically induced mutations is tedious in Streptomyces. As a means of identifying genes involved in morphological differentiation and other aspects of Streptomyces biology, we set out to develop a method for generating insertional mutants of Streptomyces coelicolor A3(2), the most intensively studied of the Streptomyces (5). Although several transposon systems have been described for use in Streptomyces (6–9), none have been used successfully to generate large numbers of random insertions, and in only two cases have insertion mutants (10, 11) been recovered in genomewide screens in which the mutant phenotype could be attributed to the transposon insertion.
As a fresh approach to the challenge of generating insertional mutants in Streptomyces on a genomewide basis, we took advantage of three recent technical advances: improvements in DNA-mediated transformation of Streptomyces (12), the emerging availability of a genome sequence for S. coelicolor (5), and the development of efficient systems for carrying out transposition in vitro. Biochemical systems now have been described for carrying out transposition in vitro with high efficiency for the transposable elements Tn7 (13), Ty1 (14), Tn5 (15), mariner (16, 17) Tn552 (18), and Mu (19). Several of these systems have been used to carry out insertional mutagenesis of bacteria that are naturally competent, such as Haemophilus influenzae (20–22) and Streptococcus pneumoniae (21).
Here we describe the introduction into S. coelicolor of plasmid libraries of S. coelicolor DNA that had been subjected to insertional mutagenesis with derivatives of mariner or Tn5. After selection for a transposon-borne drug-resistance gene, we screened for colonies in which the transposon had been incorporated into the corresponding region of homology with the chromosome by double (marker-replacement) recombination. Sequence analysis showed that transposon insertions had been recovered at scattered locations around the S. coelicolor chromosome and revealed a wide spectrum of heretofore unidentified genes. Two such insertions blocked the production of the pigmented antibiotic actinorhodin and enhanced apramycin sensitivity, respectively. Other insertions impaired the process of aerial mycelium formation, thereby identifying previously unrecognized genes that seem to play a role in morphological differentiation. One insertion, which resulted in a block in sporulation, was located in one of three adjacent genes that define three previously unrecognized families of paralogous genes in the S. coelicolor chromosome.
Materials and Methods
Strains and Growth Conditions.
S. coelicolor strains were M145 (prototrophic, SCP1- SCP2-) (23) or J1501 (hisA1 uraA1 strA1 SCP1- SCP2-) (24). Strains were grown on solid R2YE or minimal medium with 1% glucose (wt/vol) or in liquid yeast extract-malt extract (YEME) medium (23) at 30°C with, as indicated, 25 μg/ml apramycin sulfate or 200 μg/ml spectinomycin dihydrochloride (Sigma).
Transposons.
Transposon derivatives were created that harbored aac(3)IV, an apramycin resistance gene (25) (hereafter designated apr), from pOJ427 (Eli Lilly). The apr gene was cloned into a unique XbaI site located between the 3′ and 5′ inverted repeats of the transposable element mariner (Mos1) in pELHY6–0 (S. Goyard and S.M.B., unpublished work), yielding pELApr. Likewise, apr was cloned between the BamHI and XbaI sites of pMOD<MCS> (Epicentre Technologies, Madison, WI) to give pMODApr. Linear transposon DNA, Tn5apr, was liberated from pMODApr by digestion with PvuII.
Plasmid Libraries.
Plasmid libraries of S. coelicolor DNA were constructed in the vectors pSpec and pSpecoriT. To create pSpec, the Ω fragment from pHP45Ω (26), which contains the spectinomycin resistance gene aadA (hereafter designated spec), was first cloned into the HindIII site of pBluescript II SK(+) (Stratagene). The Ω-containing BssHII fragment isolated from this plasmid was ligated to the ColE1 origin [amplified by PCR from pBluescript II SK(+)], and the Ω-containing HindIII fragment was replaced by spec (amplified by PCR from HP45Ω). To create the conjugative vector pSpecoriT, the oriT sequence from pOJ446 (27) was cloned into the PstI site of pSpec. To create plasmid libraries, genomic DNA was isolated from S. coelicolor M145 (28), digested with Sau3A1, and cloned into the BamHI site of pSpec or pSpecoriT.
Insertional Mutagenesis.
Transposition reactions were carried out by using cloned libraries of S. coelicolor DNA (above) and pELApr as described by Tosi and Beverley (17) or Tn5apr as described by the manufacturer (Epicentre Technologies). DNA from the reaction mixtures was used to transform Escherichia coli with selection for transformants on LB plates containing spectinomycin and apramycin. Tn5apr gave a higher transposition efficiency (≈1 per 2,000 hybrid plasmids) and was used for most of the insertional mutagenesis experiments in this study.
The transposon-mutated hybrid plasmids were introduced into S. coelicolor by protoplast transformation or by conjugation from E. coli. Unmethylated preparations of the transposon-disrupted pSpec hybrid plasmids isolated from E. coli SCS110 (Stratagene) (0.5–2 μg) were transformed into M145 protoplasts (12, 23), plated on R2YE, and apramycin-resistant (AprR) transformants (≈103 per experiment) selected by overlay with apramycin after 16 h. Tn5-disrupted pSpecoriT hybrid plasmids were introduced into S. coelicolor M145 by conjugation (29) from E. coli strain ET12567(pUB307) transformed with the hybrid plasmids. Exconjugants were selected with apramycin (29).
The S. coelicolor AprR transformants or exconjugants were screened for mutant phenotypes. Colonies of interest were restreaked on R2YE plates containing apramycin. Next, colonies of AprR cells were replica-plated on R2YE plates containing spectinomycin to identify those that were AprR and spectinomycin-sensitive (about 5% of the total). To test for linkage between the transposon insertion and the mutation causing the mutant phenotype, genomic DNA was prepared (28), alkali-denatured (up to 25 μg; ref. 12), and used to transform protoplasts of the parent strain. Transformants were selected with apramycin and then scored for their phenotype. For about 20–25% of strains examined, the transposon insertion cotransformed with the mutant phenotype. Genomic DNA transformation also was used to transfer mariner-58 from the J1501 background in which it was isolated to M145 to give AGJ58.
Identification of S. coelicolor Sequences Flanking Transposon Insertions.
Genomic DNA from the insertion strains was digested with SacII or NotI, ligated to pBluescript II SK(+), and the ligated DNA was used to transform E. coli followed by selection with apramycin. Plasmid DNA from the AprR transformants was isolated, and flanking DNA was sequenced by using primers designed to anneal to opposite ends of apr.
Results and Discussion
Strategy for Generating Insertional Mutants.
Plasmid libraries of S. coelicolor DNA were subjected to transposition in vitro by using derivatives of the transposons Tn5 (Tn5apr) or mariner. The products of the reactions were introduced into E. coli followed by selection for both vector-borne (spec) and transposon-borne (apr) drug resistance genes, thereby enriching for transformants that harbored hybrid plasmids with a transposon insertion. Next, the transposon-bearing hybrid plasmids were introduced into S. coelicolor by protoplast transformation or by conjugation from E. coli followed by selection for AprR. Finally, we screened the AprR cells for those that were spectinomycin-sensitive. This distinguished integrants that arose from single, reciprocal (Campbell-like) recombination in which the entire transposon-bearing, hybrid plasmid had inserted into the chromosome from integrants that arose from double (marker-replacement) recombination in which only the transposon had been inserted.
Chromosomal Locations of Transposon Insertions.
We characterized 18 such AprR–spectinomycin-sensitive integrants, 17 of which were generated with Tn5apr and one with mariner. The site of insertion of each transposon was determined by sequencing DNA flanking the element. For strain AGJ58, which contains mariner, we observed the expected insertion into, and duplication of, a TA dinucleotide (base pairs 7762–7763 of SC4H8). Tn5 insertion causes duplication of a 9-bp target sequence that exhibits base preferences at certain positions (30). The Tn5apr insertions characterized were indeed flanked by a duplication of a 9-bp stretch of chromosomal DNA (Table 1). Sequences flanking the transposon for each mutant strain were used to search the Sanger Centre database (ftp://ftp.sanger.ac.uk/pub/S_coelicolor/sequences) to position the transposon on the S. coelicolor chromosome (Fig. 1). A perfect match to the database was found for sequences flanking 15 of the 18 transposon insertions, which were located at scattered sites around the chromosome.
Table 1.
Tn5apr insertion sites
| Strain | Disrupted sequence (5′→3′)* | Cosmid† |
|---|---|---|
| AG24 | 1084 CGGCACGGCCGCCGACGACGCCCG 1061 | SC8B7 |
| AG137 | 1665 CGGAGCGGCGTGAGATCCTGCGCG 1642 | SCE134 |
| AG259 | 11872 GCGCACACCGACGCCCTGTCCCGG 11895 | St9A4 |
| AG534 | 16012 GCCGACGAGCTGGGCCCGCGCGGC 16035 | SCI8 |
| AG643 | 11275 CGCGTCACCGCAGATCTGGGCGCC 11298 | SCH69 |
| AG671 | 12027 GGGTGTAGTACAGGTCGATGTGGT 12004 | SCD17 |
| AG848 | GTCGACACCTATGCTGCGGTCATC | ? |
| AG860 | 19659 GCGGACGCCTACCGGCAGACCGAG 19636 | SCF42 |
| AG1057 | CGAAGCCTGCCTCAACCTGACTCA | ? |
| AG1060 | 20511 GAGCACCGTGCTGAGGCTGGCGGC 20488 | SCAH10 |
| AG1061 | 1895 TCCCGCCCCTCAAGAGGCAGGCTC 1872 | SCH17 |
| AG1069 | TGGACCAGCAAGTGCCCATGGAGT | ? |
| AG1225 | 4955 CTTGGAACGTGCGGGTCGAACAGG 4932 | SCJ11 |
| AG1329 | 19008 GCACGCACTCTCGCCCCACCGGGC 18985 | SCF43 |
| AG1338 | 6054 GAGGACGGACACGACCCAGCGCAG 6031 | SCD25 |
| AG1346 | 28787 GGCGGCGGCACCGCTCCGGGCGCC 28810 | 2St10A7 |
| AG1359 | 15322 TGGGCAACCACCACCGCGCCGAGA 15299 | 2StG58 |
Transposon insertion sites are shown in bold along with flanking DNA sequence. Numbers denote the bp location of the sequence in the indicated S. coelicolor cosmid if known.
Cosmid sequence information is available in the European Molecular Biology Laboratory/GenBank/DNA Data Base in Japan databases (only those labeled SC) and/or from the S. coelicolor Sequencing Group at the Sanger Centre at ftp://ftp.sanger.ac.uk/pub/S_coelicolor/sequences.
Figure 1.
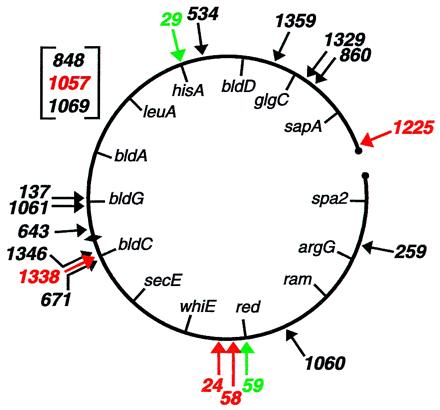
Chromosomal locations of transposon insertions. Black arrows indicate insertions that arose from double recombination and exhibited no conspicuous phenotype, red arrows indicate insertions from double recombination that were associated with a mutant phenotype, and green arrows indicate integrants in which the transposon-containing plasmid had inserted into the chromosome by single recombination. The map positions of insertions 848, 1057, and 1069 await completion of the genome sequence. Known genes are indicated inside the chromosome (31). Circles represent the ends of the linear S. coelicolor chromosome, and the diamond represents the origin of replication.
At least six insertions that did not cause a detectable phenotype were located within ORFs. These were insertions in the unknown genes SCE134.01c, SCF42.21c, and SCH69.11c, the putative oxidoreductase genes SCI8.16 and SCD17.13, and the putative ABC transporter gene SCAH10.23. Five other insertions did cause a mutant phenotype and are described below.
An Insertion Mutation Altering Pigment Production.
One of the insertion mutants, strain AG1338, exhibited a striking pigment production phenotype (Fig. 2). AG1338 failed to produce the blue pigment (actinorhodin) that is characteristic of the wild-type strain M145. The substrate mycelium of AG1338 initially exhibited a light orange color and then turned brown as growth proceeded. AG1338 was sporulation-proficient (indeed, sporulation may have been faster than in the wild type), but the spores developed a darker color than that characteristic of the wild type. S. coelicolor spores are normally gray, but whiE, which is responsible for the spore pigment, can give rise to a related brown pigment (32, 33, 46). Conceivably, AG1338 overproduces the whiE-related brown pigment at the expense of actinorhodin.
Figure 2.
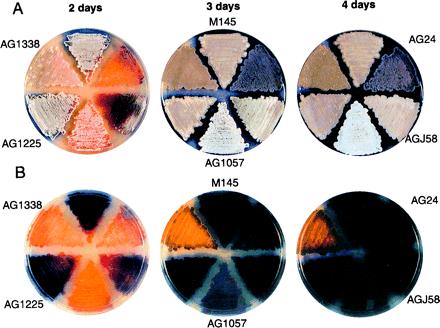
S. coelicolor insertional mutants. Shown are color photographs of colonies of the wild-type parent strain M145 and the indicated insertional mutants that had been grown on solid R2YE medium. (A) Images from the top of the plates. (B) Corresponding images from the back of the plates.
The Tn5apr insertion in AG1338 was located within a previously uncharacterized ORF, SCD25.05, which encodes a putative integral membrane protein of unknown function. Because Streptomyces is known to exhibit a significant spontaneous mutation rate (34, 35, 47), it was important to determine whether the phenotype of AG1338 was caused by the transposon insertion or by an unlinked spontaneous mutation. To investigate this, we carried out transformation with chromosomal DNA (12) to ask whether the transposon insertion would cotransform with the mutation, causing brown pigment production. Eleven of 11 of the AprR transformants exhibited a phenotype identical to that of the parent strain AG1338.
An Insertion Causing Increased Apramycin Sensitivity.
Strain AG1225 grew normally on R2YE (rich) medium (Fig. 2) with or without apramycin and on minimal medium without drug, but grew slowly on minimal medium supplemented with apramycin (25 μg/ml) (data not shown). The apramycin sensitivity mutation was shown to be 100% (43 of 43 transformants) linked to the Tn5apr insertion by cotransformation experiments with chromosomal DNA. This insertion was located to an intergenic region 36 bp downstream of gene SCJ11.08c in a large direct repeat that flanks this gene. The SCJ11.08c gene product is 98% identical to the product of a previously described S. lividans gene (ORF2) found at the junction with the terminal inverted repeat at one end of the linear chromosome of this organism (36). The product of SCJ11.08c also shows strong similarity to integral membrane transport proteins including chloramphenicol resistance protein homologs and metabolite transporters as determined by a blast search of the protein databases (37). Perhaps the SCJ11.08c gene product contributes to AprR, and the Tn5apr-1225 insertion destabilizes the SCJ11.08c transcript, reducing levels of the putative efflux transporter and thereby allowing an inhibitory concentration of apramycin to accumulate in the cell when grown on minimal medium. Conceivably and alternatively, insertion into the direct repeat adjacent to SCJ11.08c might interfere with expression of the transposon-borne apr gene.
An Insertion Causing Formation of Abnormal Aerial Hyphae.
Strain AG24 formed an abnormal aerial mycelium that was purple in color and became purplish-pink as development proceeded (Fig. 2). The aerial hyphae of AG24 were abnormally short, and the mutant colonies exhibited cracks over their surface. Light microscopy revealed that AG24 produced spores, but some were aberrantly sized (larger or smaller than the wild type), and their quantity was significantly reduced as compared with the M145 parent (data not shown). The Tn5apr insertion in AG24 was 100% (46 of 46 transformants) linked to the mutation causing abnormal aerial hyphae formation in cotransformation experiments with chromosomal DNA. The insertion was located within the ORF SC8B7.01c/SC5H4.01, the inferred product of which is annotated as containing two possible transmembrane domains and a possible ATP/GTP binding site (P-loop).
An Insertion Causing Delayed Aerial Mycelium Formation.
Mutant strain AGJ58 showed an approximately 1-day delay in aerial mycelium formation, and the aerial mycelium exhibited a pinkish-gray color (Fig. 2). Transformation experiments with chromosomal DNA showed that the mariner insertion in AGJ58 was 100% (50 of 50 transformants) linked to the mutation causing delayed aerial mycelium formation. The insertion was located in an intergenic region between the ORF SC4H8.12c and a pair of downstream, partially overlapping ORFs, SC4H8.11c and SC4H8.10c. As determined by a blast search of the protein databases (37), the inferred product of the upstream ORF SC4H8.12c exhibits strong similarity to bacterial hydroxylases. SC4H8.11c and SC4H8.10c are similar to a pair of genes, Rv1065 and Rv1066, found in Mycobacterium tuberculosis. The SC4H8.11c gene product shows weak similarity to cysteine dioxygenases, whereas the SC4H8.10c product is weakly similar to putative thiosulfate sulfurtransferases. Interestingly, the activity of a cysteine dioxygenase is believed to play a role in the transition from the mycelial to the yeast phase in the dimorphic pathogenic fungal species Histoplasma capsulatum (38–40).
An Insertion Mutation Blocking Spore Formation at an Early Stage.
Strain AG1057 exhibited a so-called white (whi) mutant phenotype (1), that is, it produced a normal-looking aerial mycelium but was markedly impaired in spore formation and the production of spore-associated gray pigment (Fig. 2). As shown in Fig. 3, at a time (4 days) when the aerial hyphae of the parent strain M145 had sporulated, the aerial hyphae of AG1057 remained undifferentiatied, appearing as straight, branched, or somewhat wavy filaments. Cotransformation experiments with chromosomal DNA demonstrated 100% (12 of 12 transformants) linkage between the Tn5apr insertion in AG1057 and the mutation causing the whi mutant phenotype.
Figure 3.
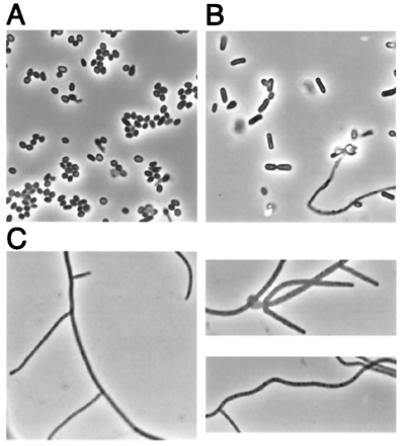
Phase-contrast micrographs of the wild-type strain M145 (A), a whiJ strain bearing the mutation whi-77 (B), and the insertional mutant AG1057 (C). Slides were prepared by taking an impression of the surface of colonies grown for 4 days on solid R2YE medium. Visualization was with an Olympus BX60 microscope at ×100 magnification.
DNA flanking the Tn5apr insertion in AG1057 was sequenced, but no match was found in the Sanger Centre database. There was, however, a perfect match to a portion of a 3.8-kb sequence that had been deposited in GenBank (accession number AF106004). The Tn5apr insertion was located within a 283-codon ORF in the deposited sequence. blast analysis (37) of this ORF failed to reveal strong similarities to sequences outside of the genus Streptomyces. Remarkably, however, 21 paralogs were detected within the current S. coelicolor database, and these were located at widely scattered sites on the chromosome (Fig. 4). The 283-codon ORF is just upstream of, and in the same orientation as, an ORF of 63 codons and downstream of an oppositely oriented ORF of 141 codons. The 63-codon ORF and the 141-codon ORF each were also represented by at least 20 paralogs at dispersed locations in the S. coelicolor genome (Fig. 4). Members of all three families of paralogs exhibit several blocks of strikingly conserved amino acid coding sequences along their lengths, examples of which are shown in Fig. 5. We also note the presence of a potential helix–turn–helix motif near the N terminus of the 283-codon ORF and each of its paralogs as revealed by Pfam analysis (41). Strikingly, 16 of the 21 paralogs of the 283-codon ORF were located immediately upstream of a paralog of the 63-codon ORF, and in eight of these 16 cases a paralog of the 141-codon ORF also was found upstream of the 283-codon paralog in the opposite orientation. We conclude that Tn5apr-1057 defines three novel families of paralogous genes and that members of each family frequently are found adjacent to each other in a characteristic arrangement.
Figure 4.
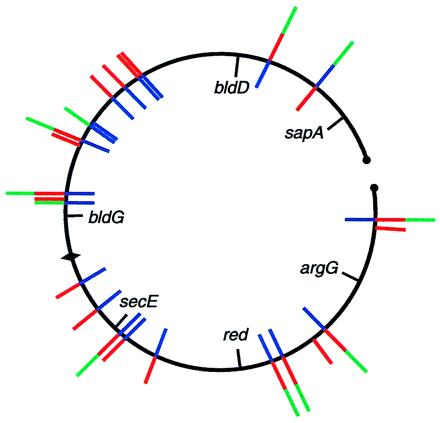
Chromosomal locations of members of the 283-codon ORF (red), 63-codon ORF (blue), and 141-codon ORF (green) families of paralogous genes. Clusters of adjacent ORFs are designated by end-to-end blue, red, and green lines. Counterclockwise from the left end of the chromosome are: SCF42.12c-14; 2StG1 base pairs 14464–13952, 14680–15540, 15550–15792; SC1G2.07c, 08c; SC1G2.14c, 15c; SC4A7.09, 10; SC121.16, 17; SCC57a.09c; SCC57a.10c, 11; SCE6.01c, 02, base pairs 1594–1800; SCE6.06; SCE94 base pairs 13886–13695, 16c, base pairs 14791–15471; SCE9.28c; SCE9.30, 31c; SCD66.13, 14; SCD6.19, 20; SCD31.02c-04; StD77 base pairs 31404–31135, 32227–31388; St9E12 base pairs 9058–8852, 9870–9055; SC9B2.15c-17; SC2H4.17c, SCAH10.01c, 02; SC5C7.22c; SC4G2.02c-04; St5F1 base pairs 32071–32946; St2H2 base pairs 12405–11959, 12579–13451, 13448–13717. ORFs designated by cosmid base pair numbers have not yet been annotated by the Sanger Centre Sequencing Group. SCF42.13, SCE9.31c, and St2H2 base pairs 13448–13717 are included as members of the 63-codon family but are more weakly similar than are other family members. Only paralogs of the 141-codon ORF that are associated with a 283-codon and/or a 63-codon family member are shown.
Figure 5.
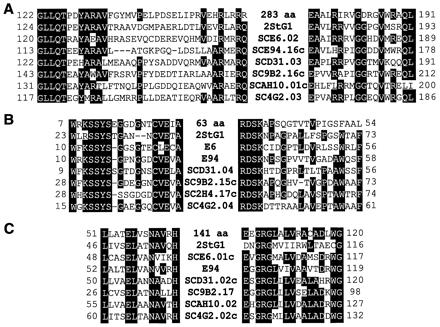
Alignment of conserved amino acid sequences for inferred protein products of 283-codon paralogs (A), 63-codon paralogs (B), and 141-codon paralogs (C). The top line in each alignment corresponds to the ORF identified by the Tn5apr-1057 insertion. The other examples correspond to cases identified in Fig. 4 in which a 283-codon family member is flanked downstream by a paralog of the 63-codon ORF and upstream by an oppositely oriented paralog of the 141-codon ORF. All eight proteins are similar along their lengths, but only a subset of the conserved regions are shown.
Because the Sanger sequencing project has not yet included the 283-codon ORF, the location of Tn5apr-1057 is not known. DNA corresponding to GenBank accession number AF106004 was assigned as the whiJ locus and was previously mapped by DNA hybridization to the overlap of cosmids D39 and D82 (31). But the sequences of D39 and D82 are now known (Sanger Centre) and evidently do not contain the AF106004 sequence. Moreover, the phenotype of the Tn5apr-1057 insertion mutant was much more severe than that of a whiJ mutant (whi-77; refs. 1 and 42) as shown in Fig. 3. Conceivably, whi-77 is a leaky allele of the 283-codon ORF. Alternatively, whi-77 could be allelic to the 63-codon ORF, the 141-codon ORF or another ORF in the 3.8-kb segment of DNA. It will be interesting to see where Tn5apr-1057 (and whiJ) is located on the S. coelicolor chromosome once the Sanger sequencing project is completed.
Insertional Mutants Arising from Single Recombination.
Transposon integration also can arise by single, reciprocal recombination in which the entire transposon-bearing, hybrid plasmid is incorporated into the chromosome. Such AprR–spectinomycin-resistant integrants could exhibit a mutant phenotype if the S. coelicolor insert carried by the plasmid is internal to a transcription unit (43). Two mutant strains that arose from integration of a hybrid plasmid were AG29 and AG59. Strain AG29 was auxotrophic and strain AG59 failed to produce the red pigment undecylprodigiosin. In both cases, the vector-borne drug resistance gene was 100% linked to the mutation responsible for the mutant phenotype in cotransformation experiments with chromosomal DNA. DNA sequence analysis showed that AG29 harbored a hybrid plasmid insert (base pairs 13638–18321 of cosmid SC4G6; Fig. 1) within the histidine biosynthesis operon (44). Consistent with this, the mutant was able to grow on minimal medium that had been supplemented with histidine (not shown). Likewise, AG59 harbored a hybrid plasmid insert within a cluster of (red) genes (base pairs 17370–22882 of cosmid SC3F7; Fig. 1) responsible for undecylprodigiosin biosynthesis (31, 45).
Summary.
We have succeeded in introducing insertions of the transposons Tn5 or mariner at widely scattered locations around the S. coelicolor chromosome. This has revealed multiple novel genes associated with defects in secondary metabolism, drug resistance, and morphological differentiation. In each case cotransformation with chromosomal DNA showed that the transposon insertion was linked to the mutation causing the mutant phenotype. Specifically, we identified an insertion in a previously uncharacterized gene that blocked actinorhodin production and resulted instead in the production of a brown pigment. We identified an insertion that caused apramycin sensitivity and is located adjacent to a gene whose inferred product is similar to antibiotic efflux pumps. We recovered two insertions in or near uncharacterized genes that delayed or impaired aerial mycelium formation. We also identified an insertion that blocked the conversion of aerial hyphae into chains of spores, defining a previously uncharacterized whi gene. Interestingly, the whi gene and two flanking ORFs are each members of novel families of paralogous genes. Individual members from each family frequently are found clustered together in a characteristic arrangement at dispersed locations in the chromosome. It will be interesting to determine whether the three gene families have related functions and whether any of the additional gene clusters are involved in morphological differentiation. The stage is now set to use insertional mutagenesis on a large scale for discovering genes involved in morphological differentiation, secondary metabolism, and other aspects of Streptomyces biology.
Acknowledgments
We thank S. Goyard for pELHY6–0, Z. Li for pELApr, and S. Goyard and J. Guzova for transposition tests of pELApr. We thank M. Buttner, K. Chater, D. Hopwood, J. McCormick, L. Shapiro, and J. Westpheling for helpful comments on the manuscript. This work was supported by a grant (to R.L.) from the National Science Foundation (MCB-9727234) and National Institutes of Health Grant AI29646 (to S.M.B.). A.M.G. is supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship, DRG-1524.
Abbreviation
- AprR
apramycin-resistant
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170059797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170059797
References
- 1.Chater K F. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 2.Kelemen G H, Buttner M J. Curr Opin Microbiol. 1998;1:656–662. doi: 10.1016/s1369-5274(98)80111-2. [DOI] [PubMed] [Google Scholar]
- 3.Champness W. In: Prokaryotic Development. Brun Y V, Shimkets L J, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 11–31. [Google Scholar]
- 4.Bibb M. Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- 5.Hopwood D A. Microbiology. 1999;145:2183–2202. doi: 10.1099/00221287-145-9-2183. [DOI] [PubMed] [Google Scholar]
- 6.Olson E R, Chung S-T. J Bacteriol. 1988;170:1955–1957. doi: 10.1128/jb.170.4.1955-1957.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baltz R H, Hahn D R, McHenney M A, Solenberg P J. Gene. 1992;115:61–65. doi: 10.1016/0378-1119(92)90541-v. [DOI] [PubMed] [Google Scholar]
- 8.Volff J-N, Altenbuchner J. Gene. 1997;194:81–86. doi: 10.1016/s0378-1119(97)00163-7. [DOI] [PubMed] [Google Scholar]
- 9.Herron P R, Evans M C, Dyson P J. FEMS Microbiol Lett. 1999;171:215–221. doi: 10.1111/j.1574-6968.1999.tb13435.x. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda H, Takada Y, Pang C-H, Tanaka H, Omura S. J Bacteriol. 1993;175:2077–2082. doi: 10.1128/jb.175.7.2077-2082.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McHenney M A, Hosted T J, Dehoff B S, Rosteck P R, Jr, Baltz R H. J Bacteriol. 1998;180:143–151. doi: 10.1128/jb.180.1.143-151.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh S-H, Chater K F. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bainton R J, Kubo K M, Feng J, Craig N L. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- 14.Devine S E, Boeke J D. Nucleic Acids Res. 1994;22:3765–3772. doi: 10.1093/nar/22.18.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goryshin I Y, Reznikoff W S. J Biol Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 16.Lampe D J, Grant T E, Robertson H M. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tosi L R O, Beverley S M. Nucleic Acids Res. 2000;28:784–790. doi: 10.1093/nar/28.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin T J, IV, Parsons L, Leschziner A E, DeVost J, Derbyshire K M, Grindley N D F. Nucleic Acids Res. 1999;27:3859–3865. doi: 10.1093/nar/27.19.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haapa S, Taira S, Heikkinen E, Savilahti H. Nucleic Acids Res. 1999;27:2777–2784. doi: 10.1093/nar/27.13.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gwinn M L, Stellwagen A E, Craig N L, Tomb J-F, Smith H O. J Bacteriol. 1997;179:7315–7320. doi: 10.1128/jb.179.23.7315-7320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reich K A, Chovan L, Hessler P. J Bacteriol. 1999;181:4961–4968. doi: 10.1128/jb.181.16.4961-4968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic Manipulation of Streptomyces: A Laboratory Manual. Norwich, U.K.: The John Innes Foundation; 1985. [Google Scholar]
- 24.Chater K F, Bruton C J, King A A, Suarez J E. Gene. 1982;19:21–32. doi: 10.1016/0378-1119(82)90185-8. [DOI] [PubMed] [Google Scholar]
- 25.Bräu B, Pilz U, Piepersberg W. Mol Gen Genet. 1984;193:179–187. doi: 10.1007/BF00327434. [DOI] [PubMed] [Google Scholar]
- 26.Prentki P, Krisch H M. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 27.Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 28.Pospiech A, Neumann B. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 29.Flett F, Mersinias V, Smith C P. FEMS Microbiol Lett. 1997;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 30.Goryshin I Y, Miller J A, Kil Y V, Lanzov V A, Reznikoff W S. Proc Natl Acad Sci USA. 1998;95:10716–10721. doi: 10.1073/pnas.95.18.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 32.Davis N K, Chater K F. Mol Microbiol. 1990;4:1679–1691. doi: 10.1111/j.1365-2958.1990.tb00545.x. [DOI] [PubMed] [Google Scholar]
- 33.Horinouchi S, Beppu T. J Bacteriol. 1985;162:406–412. doi: 10.1128/jb.162.1.406-412.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leblond P, Decaris B. FEMS Microbiol Lett. 1994;123:225–232. doi: 10.1111/j.1574-6968.1994.tb07229.x. [DOI] [PubMed] [Google Scholar]
- 35.Volff J-N, Altenbuchner J. Mol Microbiol. 1998;27:239–246. doi: 10.1046/j.1365-2958.1998.00652.x. [DOI] [PubMed] [Google Scholar]
- 36.Volff J-N, Viell P, Altenbuchner J. Mol Gen Genet. 1997;253:761–765. doi: 10.1007/s004380050381. [DOI] [PubMed] [Google Scholar]
- 37.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maresca B, Lambowitz A M, Kumar V B, Grant G A, Kobayashi G S, Medoff G. Proc Natl Acad Sci USA. 1981;78:4596–4600. doi: 10.1073/pnas.78.7.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sacco M, Maresca B, Kumar B V, Kobayashi G S, Medoff G. J Bacteriol. 1981;146:117–120. doi: 10.1128/jb.146.1.117-120.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar V, Maresca B, Sacco M, Goewert R, Kobayashi G S, Medoff G. Biochemistry. 1983;22:762–768. doi: 10.1021/bi00273a009. [DOI] [PubMed] [Google Scholar]
- 41.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L L. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chater K F, Merrick M J. In: Second International Symposium on the Genetics of Industrial Microorganisms. MacDonald K D, editor. London: Academic; 1976. pp. 583–593. [Google Scholar]
- 43.Chater K F, Bruton C J. Gene. 1983;26:67–78. doi: 10.1016/0378-1119(83)90037-9. [DOI] [PubMed] [Google Scholar]
- 44.Limauro D, Avitabile A, Cappellano C, Puglia A M, Bruni C B. Gene. 1990;90:31–41. doi: 10.1016/0378-1119(90)90436-u. [DOI] [PubMed] [Google Scholar]
- 45.Malpartida F, Niemi J, Navarrete R, Hopwood D A. Gene. 1990;93:91–99. doi: 10.1016/0378-1119(90)90141-d. [DOI] [PubMed] [Google Scholar]
- 46.Yu T-W, Hopwood D A. Microbiology. 1995;141:2779–2791. doi: 10.1099/13500872-141-11-2779. [DOI] [PubMed] [Google Scholar]
- 47.Nodwell J R, McGovern K, Losick R. Mol Microbiol. 1996;22:881–893. doi: 10.1046/j.1365-2958.1996.01540.x. [DOI] [PubMed] [Google Scholar]


