Abstract
Gene duplication is one of the most important mechanisms for creating new genes and generating genomic novelty. Retrotransposon-mediated sequence transduction (i.e., the process by which a retrotransposon carries flanking sequence during its mobilization) has been proposed as a gene duplication mechanism. L1 exon shuffling potential has been reported in cell culture assays, and two potential L1-mediated exon shuffling events have been identified in the genome. SVA is the youngest retrotransposon family in primates and is capable of 3′ flanking sequence transduction during retrotransposition. In this study, we examined all of the full-length SVA elements in the human genome to assess the frequency and impact of SVA-mediated 3′ sequence transduction. Our results showed that ≈53 kb of genomic sequences have been duplicated by 143 different SVA-mediated transduction events. In particular, we identified one group of SVA elements that duplicated the entire AMAC gene three times in the human genome through SVA-mediated transduction events, which happened before the divergence of humans and African great apes. In addition to the original AMAC gene, the three transduced AMAC copies contain intact ORFs in the human genome, and at least two are actively transcribed in different human tissues. The duplication of entire genes and the creation of previously undescribed gene families through retrotransposon-mediated sequence transduction represent an important mechanism by which mobile elements impact their host genomes.
Keywords: gene duplication, SVA, retrotransposition, mobile element
The emergence of new genes and biological functions is crucial to the evolution of species (1). Several mechanisms for creating new genes are known (1), the best characterized pathway being through duplication of preexisting genes (1, 2). Several types of duplications leading to genetic innovation have been investigated, including segmental duplication (3) and gene retrotransposition (4–7). Here, we investigate a less well characterized mechanism that can potentially duplicate genes, namely the transduction of flanking genomic sequence associated with the retrotransposition of mobile elements.
Retrotransposons usually do not carry downstream motifs that are important for efficient transcription termination. Therefore, when they are transcribed, the RNA transcription machinery sometimes skips the element's own weak polyadenylation signal and terminates transcription by using a downstream polyadenylation site located in the 3′ flanking genomic sequence. The transcript containing the retrotransposon along with the extra genomic sequence is subsequently integrated back into the genome through retrotransposition, a process termed 3′ transduction (8, 9). In principle, this mechanism could lead to the duplication of coding sequences located in the transduced flanking genomic sequence. Indeed, the exon shuffling and genetic diversity of the L1-mediated 3′ transduction has been demonstrated in cell culture assays (9, 10). However, among all of the studies investigating L1-mediated exon shuffling (11–16), only two putative examples of exon transduction have been reported in the human genome (15, 16).
The SVA family of retrotransposons originated <25 million years ago (mya) and has increased to ≈3,000 copies in the human genome (17, 18). Similar to L1 elements, SVA elements are thought to be transcribed by RNA polymerase II and have the ability to transduce downstream sequence (18). Approximately 10% of human SVA elements appear to have been involved in sequence transduction events (17). Here, we examined the extent and properties of SVA-mediated transduction events to evaluate their evolutionary impact on the human genome. Our results demonstrate that retrotransposon-mediated sequence transduction is not only a mechanism for exon shuffling but also serves as a previously uncharacterized mechanism for gene duplication and the creation of new gene families.
Results and Discussion
Genomic Analysis for SVA 3′ Transductions.
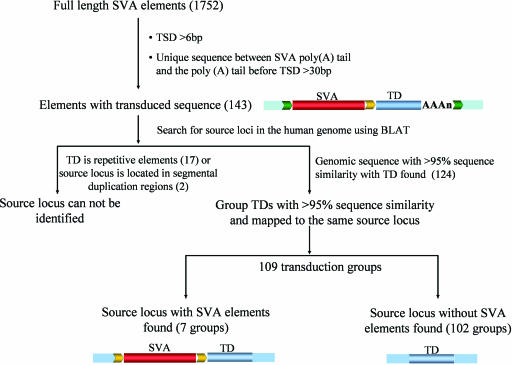
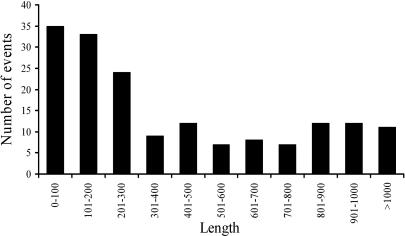
To investigate the extent of transduction events associated with SVA elements, we examined all 1,752 full-length SVA elements in the human genome reference sequence. In total, 143 SVA elements with putative transduced sequences were identified according to our validation criteria (Fig. 1; see Materials and Methods for details). Most of these loci (123 of 143) also displayed typical AATAAA or ATTAAA polyadenylation signals located 5–52 nt upstream of the start of the poly(A) tail, further supporting that these loci represent authentic SVA-mediated transduction events. The size of the transduced sequences ranged from 35 to 1,853 bp, with an average of 340 bp (Fig. 2). Overall, 52,740 bp of genomic sequences were duplicated by these SVA-mediated transductions. To determine whether the transduction events are specific characteristics of particular SVA subfamilies, SVA elements with transduced sequence were aligned and compared with all known SVA subfamily consensus sequences (17). We found transduction events involving all previously identified SVA families, suggesting that 3′ transduction is a common phenomenon among SVA members (see Table 1, which is published as supporting information on the PNAS web site). Given that full-length SVA elements comprise 63% of the family, we extrapolate that SVA elements may have transduced a total of ≈84 kb of genomic sequence during their expansion.
Fig. 1.
Identification of SVA 3′ transduction events and their source elements. Shown are the schematic diagrams for the identification process. Flanking sequences of the source locus are shown as blue boxes; TSDs are shown as yellow and green arrows. SVA elements are depicted as red bars, and the transduced sequences are shown as blue bars and labeled “TD.” SVA element poly(A) tails are shown as “(AAA)n.” The numbers in parentheses correspond to the total number of SVA elements/groups identified in each step.
Fig. 2.
Length distribution of 3′ transduction events. The number of human SVA-mediated 3′ transduction events in each 100-bp size interval is shown.
The rate of SVA-mediated transduction events (8.2%) is similar to the L1 transduction rates reported in previous studies (11, 12, 14). Our results are likely to be an underestimate because the method we used to validate candidate transductions relied on the detection of perfect target site duplications (TSDs). This requirement may miss transduction events as a result of substitutions in their TSDs. Detailed information concerning all of the SVA transduction events reported in this article can be found in Table 2, which is published as supporting information on the PNAS web site.
Identification of Transduction Source Loci.
To identify the source loci of the transduced sequences, we searched the human genome by using the BLAST-like alignment tool (BLAT) (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start). With the exception of 19 loci in which the transduced sequence was totally composed of repetitive sequences or source loci were located in segmental duplications (Fig. 1) and, thus, could not be mapped precisely to any location in the human genome, we were able to identify the source loci for each of the other 124 transductions. Although 98 transduced loci could be uniquely linked to their source locus and each of them was treated as a unique group, the remaining 26 transduced loci showed >95% sequence identity with at least one other transduced locus and were mapped to the same source locus. Hence, the 26 transduced loci were assigned to 11 transduction groups, each of which contained two or more transduced loci.
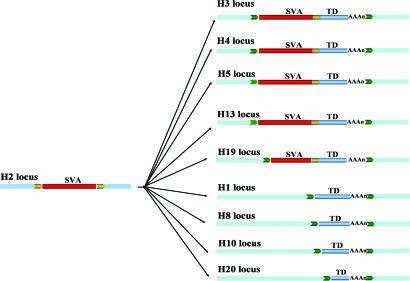
Seven of the total 109 source loci contained an SVA element (Fig. 1). These source SVA elements could be unambiguously identified because only the SVA elements were surrounded by TSDs. By comparison, in the transduced loci, the TSDs included both the SVA element and the transduced flanking genomic sequence. The source SVA element for the transduction group H3_186 located on human chromosome 2p11.2 represented one of the most active elements we identified, given that it generated at least nine transduced copies (Fig. 3). Sequence comparisons showed that the source locus and all nine transduced loci were absent from the chimpanzee genome. This result suggests that the source locus inserted in the human genome after the human–chimpanzee divergence and generated all of the loci with transductions within the last ≈6 million years. Among the nine transduced loci, five were typical SVA transduction loci (i.e., having both the SVA element and the transduced sequence), whereas four loci contained only the transduced genomic sequences. Although there is no SVA element upstream of these sequences, a poly(A) tail can be found downstream of the transduced sequence, and the TSDs surrounding the transduced sequence are identifiable. Presumably, these loci were generated via SVA-mediated transduction events associated with 5′ truncation during the integration process or through incomplete reverse transcription during retrotransposition. SVA represents one of the most active retrotransposon families in humans (19), and little is known regarding their retrotransposition mechanism. Because the source loci with SVA elements may be capable of retrotransposition, detailed analysis of the seven transduction source loci we identified in this study may shed light on the underlying mechanism of SVA retrotransposition.
Fig. 3.
SVA 3′ transduction events. One group of SVA 3′ transduction events (H3_186) is shown. Flanking sequences of the original locus are shown as blue boxes, and the flanking sequences of the transduced loci are shown as light blue boxes. TSDs are shown as yellow and green arrows. SVA elements are depicted as red bars, and the transduced sequences are shown as blue bars and labeled “TD.” SVA element poly(A) tails are shown as “(AAA)n.”
For the other 102 source loci, only sequences corresponding to transduced sequences were present. Each of the loci was devoid of the SVA elements, TSDs, and poly(A) tails. The simplest explanation for this genomic configuration is that the retrotransposition events carrying transduced sequence occurred while the source SVA element was polymorphic for insertion presence/absence in the population. Subsequently, the original locus with the source SVA element may have been lost in the population or still may be polymorphic in the human population but absent from the human genome reference sequence. To test source loci for insertion polymorphism, we genotyped 30 source loci in 80 diverse human genomes by using PCR assays as described in ref. 17. No source loci containing an SVA element were present in any of the diverse human genomes that were surveyed for the 30 SVA source elements. These results suggest that most of the SVA elements that generated transduction events were subsequently lost from the human genome. This observation is not surprising because similar results have been observed in L1-mediated transduction studies (14, 20). In fact, the majority of new mobile element insertions are expected to be lost from the population because of genetic drift under neutral evolution. A second potential reason for the disappearance of these source elements may be moderate negative selection as a result of their transcription and retrotransposition capacity (20).
SVA Transduction-Mediated Gene Duplication.
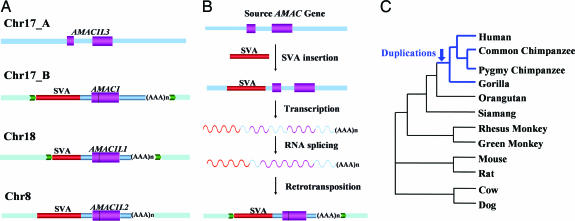
One particularly interesting example of SVA-mediated 3′ transduction is the H17_76 group (Fig. 4A), in which four related loci were identified in the human genome. The source locus (chr17_A) contained only the transduced sequence without the SVA element, and the other three loci (chr17_B, chr18, and chr8) contained an SVA element along with the transduced sequence resulting from retrotransposition. This group of transduced sequences is among the longest of all of the elements we recovered (1,682, 1,245, and 1,257 bp for loci chr17_B, chr18, and chr8 respectively). Subsequent analysis of the transduced sequences resulted in the identification of the gene AMAC1 (acyl-malonyl condensing enzyme 1, Entrez Gene ID: 146861) at the chr17_B locus, AMAC1L1 (AMAC1-like 1, Entrez Gene ID: 492318) at the chr18 locus, AMAC1L2 (AMAC1-like 2, Entrez Gene ID: 83650) at the chr8 locus, and AMAC1L3 (AMAC1-like 3, Entrez Gene ID: 404029) at the chr17_A locus. Interestingly, the source locus (chr17_A) contains 467 bp of extra sequence in the middle of the transduced sequence as compared with the other copies. Examination of the gene structure showed that AMAC1L3 at the inferred source locus had two exons separated by an intron (the extra DNA sequence), whereas the three SVA transduced loci contained intronless versions of AMAC1L3 (Fig. 4A). These results suggest that the intron was spliced out during the retrotransposition process, providing further evidence for the underlying mechanism that created the three duplicated copies (Fig. 4B). Although the exact function of human AMAC genes has not been determined, studies in bacteria showed that AMAC is an enzyme involved in fatty acid synthesis, in which it condenses a two-carbon unit from malonyl-(acyl carrier protein) to fatty acyl-(acyl carrier protein), adding two carbons to the fatty acid chain with the release of the carbon dioxide (21).
Fig. 4.
SVA transduction-mediated gene duplication. (A) Schematic diagram of the H17_76 transduction group in the human genome. Flanking sequences of the original locus are shown as blue boxes, and the flanking sequences of the transduced loci are shown as light blue boxes. TSDs are shown as yellow and green arrows. SVA elements are depicted as red bars, the transduced sequences are shown as blue bars, and coding regions are shown as purple bars. SVA element poly(A) tails are shown as “(AAA)n.” (B) Schematic diagrams for putative evolutionary scenarios of the SVA transduction-mediated gene duplications. Approximately 7 million to 14 million years ago, one active SVA element was inserted upstream of the original AMAC gene locus. Then, transcription of this active SVA element transduced the full-length AMAC gene sequence. During the retrotransposition process, the intron of the gene was removed by RNA processing machinery. Finally, the SVA element along with the intronless AMAC gene sequence retrotransposed into new genomic locations. The original retrotransposition-competent SVA element upstream of the source locus was eventually lost in the population. The predicted RNA transcripts are shown as curved lines. (C) The phylogenetic relationships among various species used in dN/dS analysis.
Evolutionary Analyses of AMAC Genes.
To determine the evolutionary history of AMAC transduction events, we first used BLAT to search for orthologous loci corresponding to all four human AMAC genes in four available mammalian genome sequences (mouse, rat, cow, and dog). The search resulted in the identification of a single AMAC locus in all four species examined, and it was orthologous to the human AMAC1L3. In particular, the mouse orthologous region is annotated as the gene AMAC1 (Entrez GeneID: 56293). The murine AMAC1 gene contains two exons similar to human AMAC1L3 gene. Only the human AMAC1L3 locus was colinear with the mouse AMAC1 genomic sequence, whereas the three SVA transduced loci were colinear with the mouse AMAC1 mRNA sequence.
Next, we investigated the origin of the multiple AMAC gene copies within the primate lineage by analyzing seven nonhuman primate species (i.e., pygmy chimpanzee, common chimpanzee, gorilla, orangutan, siamang, African green monkey, and rhesus monkey). PCR and DNA sequence analyses showed that the intron-containing locus AMAC1L3 is present in all species examined. By contrast, the AMAC1 and AMAC1L2 loci were present only in human, pygmy chimpanzee, common chimpanzee, and gorilla genomes. Because of the presence of repetitive elements in the flanking regions, the AMAC1L1 locus could not be successfully amplified by using PCR and was excluded from subsequent analyses. Together, our results suggest that the transduction events happened after the divergence of African apes from orangutans but before the divergence of humans, chimpanzees, and gorillas, ≈7 to 14 million years ago based on the estimated divergence time of primates (22).
To determine the functional status of the AMAC genes, we first examined all available AMAC copies for intact ORFs. Examination of the DNA sequences showed that the AMAC1 loci in both common chimpanzee and gorilla contained a premature stop codon in their ORF regions, located at amino acid positions 29 and 32, respectively. Therefore, these two copies have lost their coding capacity and have become processed pseudogenes. The ORFs of all of the other AMAC copies, including all four human copies, remained intact.
Next, we examined the selective constraints on all AMAC copies by using the maximum likelihood-based program PAML (23). PAML estimates the nonsynonymous (dN) and synonymous (dS) substitution rate ratios (ω = dN/dS) as measures of selective pressure, where the value of ω indicates the type of selection (<1, purifying; =1, neutral; >1, positive). Likelihood ratio tests then can be used to compare the different models of evolution. First, a maximum-likelihood tree was constructed by using all available AMAC ORF sequences (see Materials and Methods), and ω values were estimated according to a model of a single ω among branches of the tree (Model 0) and a model where ω is allowed to vary among branches (Model 1). The results showed that Model 1 fit the data significantly better than Model 0 (P < 0.0001; see Table 3, which is published as supporting information on the PNAS web site), suggesting that different selective pressures are acting on the various AMAC copies within and between species. We then separated the branches of the tree into two groups, corresponding to the sequences predating the gene duplication events (Fig. 4C, black branches) and sequences postdating duplication events (Fig. 4C, bold blue branches), and estimated ω values for the two groups separately. The ω in the branches predating the duplications was estimated to be 0.13 (significantly different from ω = 1, P < 0.0001), suggesting that the AMAC gene is under strong purifying selection in the species lacking duplicated copies. By contrast, the ω was estimated to be 1.24 (not significantly different from ω = 1, P = 0.25) in the species that possess multiple copies of AMAC. In addition, the comparison between Model 0 and Model 1 for branches after the duplications showed that ω values among branches are not significantly different (P = 0.69). Together, these results indicate that the AMAC gene was under purifying selection in all of the species before the duplication events and that, after the gene duplication events, all AMAC copies experienced a relaxation of selective constraints.
These observations are in good agreement with the predictions of classical gene duplication theory (2, 24), which suggests that the functional redundancy of newly duplicated genes will result initially in free evolution of all gene copies. The long-term evolutionary fate of the new gene copies includes loss of function (nonfunctionalization), evolution of a new function (neofunctionalization), or maintenance of the duplicated copies for the original function (subfunctionalization). The stop codon present in different positions in the chimpanzee and gorilla AMAC1 copies shows the nonfunctionalization of these particular gene copies. Furthermore, we also analyzed the AMAC gene sequences for possible sites or domains that are (or were) under positive selection and developed new functions (neofunctionalization) (Table 3 and also Table 4, which is published as supporting information on the PNAS web site). However, the paucity of functional studies on the human AMAC genes prevented detailed validation and comparisons of the potential functional role of candidate sites.
AMAC Expression Studies.
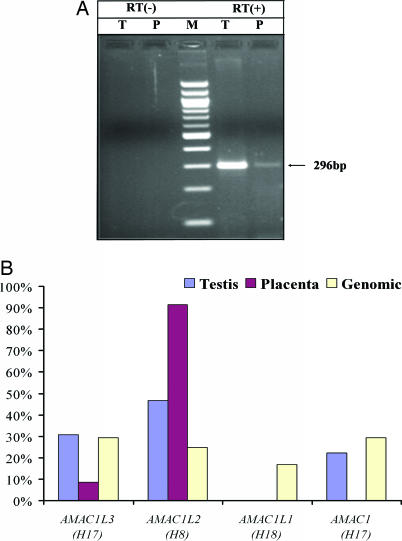
To further investigate the functional status of human AMAC gene duplicates, we examined the expression pattern of the four human AMAC gene copies. RT-PCR analysis was performed by using poly(A)-selected RNA from human testis and placenta and oligonucleotide primers designed to match conserved regions in all four gene copies (see Fig. 6, which is published as supporting information on the PNAS web site). We first amplified the target sequences from human genomic DNA (HeLa), cloned the PCR products, and sequenced ≈100 clones. Based on nucleotide substitutions specific for each AMAC gene duplicate, we determined the origin of each clone. Our results showed that all four gene copies were recovered in a comparable manner, showing that our approach amplifies the four human AMAC copies with similar efficiency (Fig. 5B). Using the same primer set, RT-PCR generated a product with the expected size in both tissues (Fig. 5A). We cloned the RT-PCR products and sequenced ≈100 clones derived from each tissue (Fig. 5B). Sequence analysis showed that AMAC1, AMAC1L2, and AMAC1L3 were expressed in testis, whereas only AMAC1L2 and AMAC1L3 were expressed in placenta. In both tissues, the SVA-transduced AMAC1L2 was predominantly expressed. Together, these results show that at least two SVA-transduced AMAC gene duplicates are expressed in humans and that they may have differential tissue expression patterns.
Fig. 5.
Expression analysis of AMAC gene duplicates in humans. (A) Agarose gel chromatograph of RT-PCR products derived from human testis (T) and placental (P) RNA templates. Negative controls with no reverse transcriptase (RT −) are on the left, a 100-bp marker (M) is in the middle, and reactions with reverse transcriptase (RT +) are on the right; the sizes of the correct fragments are indicated. (B) Relative expression levels of four human AMAC gene duplicates in human testis and placenta. Human genomic DNA (HeLa) amplification is the control for uniform amplification of all gene duplicates.
To determine the expression pattern of AMAC gene copies in human tissues, we searched the National Center for Biotechnology Information EST database for AMAC-related transcripts. Consistent with the RT-PCR results, two of the SVA-transduced AMAC copies (AMAC1 and AMAC1L2) were recovered from the EST database, and their full-length cDNAs already have been sequenced (GenBank accession nos. AK097473 and AJ291677 for AMAC1 and AMAC1L2, respectively). Both mRNA sequences start downstream from the SVA elements, suggesting that their promoters have been duplicated along with the gene copies themselves. By contrast, the two transcripts contained unique 3′ UTR sequences specific to their new genomic locations. Thus, it appears that these two gene copies have acquired new downstream polyadenylation signals subsequent to their integration in the genome.
Concluding Remarks
Our results represent an example in the primate lineage of gene duplications derived from SVA-mediated 3′ transduction. One factor that may increase the potency of SVA elements with regard to the generation of new genes is that, in addition to duplicating genes, SVA elements also may provide promoters for newly integrated gene duplicates. SVA elements contain an LTR-derived region that is used as promoter in endogenous retroviruses, and several studies have shown that LTRs in the genome can function as promoters for downstream genes (25, 26). Therefore, the SVA LTR-derived tail region might function as an alternative promoter for genes involved in 3′ transduction events whenever the original gene-specific promoter was not transduced.
Retrotransposon-mediated 3′ transduction represents a previously uncharacterized mechanism for entire gene duplication that can lead to the rapid generation of new gene families. Although the gene duplications by retrotransposon-mediated transduction reported here created intronless gene copies similar to duplications resulting from gene retrotransposition, there is one major difference between these two duplication mechanisms. Gene retrotransposition generally does not carry the promoter and regulatory region of the retrotransposed gene to its new location because of the process by which the gene is reverse transcribed into cDNA. Thus, the newly transposed gene must acquire new regulatory sequences to be functional. By contrast, retrotransposon-mediated 3′ transduction events not only duplicate whole genes, but also can duplicate promoter regions (as demonstrated by the AMAC gene duplicates). Consequently, duplications resulting from 3′ transduction retain their functional potential after inserting into their new genomic locations, allowing immediate release of the gene copies (the original and duplicates) from selective constraints.
Mobile elements already are known for creating genomic novelty in a variety of ways. For example, L1 elements provide the molecular machinery necessary for gene retrotransposition (27, 28), and new fusion genes can be generated during the process (6, 7). DNA transposons also can mediate exon shuffling and gene duplication, as demonstrated by mutator-like elements and helitron-like elements in plants (29, 30). Further, mobile elements themselves can serve as raw material for the generation of new functions by their incorporation into existing genes (31–33). By serving as a mechanism for gene duplication and generating new gene families, retrotransposon-mediated sequence transduction represents an important mechanism by which mobile elements impact their host genomes.
Materials and Methods
Genome Analysis.
The RepeatMasker annotations of the human genome reference sequence (hg17, May 2004) were obtained from the University of California, Santa Cruz Genome Bioinformatics Site (http://genome.ucsc.edu). The full-length SVA elements with a 2,000-bp flanking sequence on each side were exacted by using a perl script, and the TSDs were identified manually. SVA elements were considered as candidates for containing 3′ transduction sequence if they had (i) unambiguous TSDs (>6 bp) and (ii) a >30-bp sequence between the end of the SVA sequence and the poly(A) tail immediately upstream of the 3′ TSD. The extra sequence then was used for a sequence similarity BLAT search against the human genomic database. For genomic loci exhibiting >95% identity to the putative transduced sequence, a 5-kb extra sequence was extracted on both ends of the locus, and a locally installed RepeatMasker program was used to determine their repeat content. In some cases, segmental duplications may result in a similar pattern as the 3′ transduction. To remove such false-positive hits, we further compared the sequence directly flanking the previously uncharacterized locus with the flanking sequence of the query locus. If the flanking sequences from both loci showed >90% sequence similarity for >2 kb in length, the previously uncharacterized locus was excluded from the further analysis.
PCR/RT-PCR and DNA Sequence Analysis.
The human population panel used in the polymorphism analysis is described in ref. 17. Other DNA samples used in this study included human genomic DNA (HeLa cell line ATCC CCL-2) and the following nonhuman primate species available as a primate phylogenetic panel PRP00001 from Coriell (Camden, NJ): DNA samples of Pan troglodytes (common chimpanzee), P. paniscus (bonobo or pygmy chimpanzee), Gorilla gorilla (western lowland gorilla), Pongo pygmaeus (orangutan), and Macaca mulatta (rhesus monkey). DNA samples of Hylobates syndactylus (siamang) also were purchased from Coriell (PR00721). DNA samples of Chlorocebus aethiops (green monkey) were isolated from cell line ATCC CCL70.
The primers and annealing temperatures for each locus are shown in Table 5, which is published as supporting information on the PNAS web site, and are also available upon request. For the AMAC gene analysis, AMAC gene-related loci were amplified by using PCR with different primates as templates with different primer sets and annealing temperatures (see Table 6, which is published as supporting information on the PNAS web site).
For RT-PCR, 1 μg of poly(A)-selected RNA (Ambion, Austin, TX) from human testis and placenta was used to perform the reverse transcription reaction by using the Reverse Transcription System kit (Promega, Madison, WI) with conserved primer 4(−) (5′-CAGATAGGAAGGCCACTGTTG-3′) according to manufacturer's protocol. After completion, the volume of the reverse transcription reaction was brought to a final volume of 100 μl with nuclease-free water. Ten microliters of the reverse transcription reaction was used for PCR with conserved primer 1(+) (5′-ATTGCCCTGCTACTTAAACTGC-3′)/conserved primer 1(−) (5′-TGTAGTGTCCAGAGTCCAGGTC-3′) for 32 cycles at annealing temperature of 60°C. PCR products were fractionated on a 1.5% low melting agarose gel and extracted with the QIAquick Gel Extraction kit (Qiagen, Valencia, CA).
For sequencing analysis, individual RT-PCR/PCR products were cloned by using the TOPO-TA cloning kit (Invitrogen, Carlsbad, Ca) and sequenced by using chain termination sequencing on an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). All sequences were deposited in GenBank under accession numbers DQ482900–DQ482914.
Evolutionary Analysis.
All available AMAC gene-coding region homologous sequences were aligned by using ClustalX (34), followed by manual adjustments. Next, a maximum-likelihood tree were constructed under HKY85+G model by using PAUP* (35) The resulting tree (see Fig. 7, which is published as supporting information on the PNAS web site) was used for the dN/dS ratio analysis.
Supplementary Material
Acknowledgments
We thank everybody in the M.A.B. laboratory for critical reading and discussion during the preparation of the manuscript, in particular Dr. Scott Herke for his contribution during the revision of this manuscript, and anonymous reviewers for their useful comments on earlier versions of the manuscript. This work was supported by National Science Foundation Grants BCS-0218338 (to M.A.B.) and EPS-0346411 (to M.A.B. and P.L.D.), Louisiana Board of Regents Millennium Trust Health Excellence Fund HEF 2001-06-02 (to M.A.B.), National Institutes of Health Grants R01GM59290 (to M.A.B.) and R01GM45668 (to P.L.D.), and the State of Louisiana Board of Regents Support Fund (M.A.B.).
Abbreviation
- TSD
target site duplication.
Footnotes
References
- 1.Long M, Betran E, Thornton K, Wang W. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 2.Ohno S. Evolution by Gene Duplication. Heidelberg: Springer; 1970. [Google Scholar]
- 3.Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 4.Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinckenbosch N, Dupanloup I, Kaessmann H. Proc Natl Acad Sci USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Nature. 2004;430:569–573. [Google Scholar]
- 7.Nisole S, Lynch C, Stoye JP, Yap MW. Proc Natl Acad Sci USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH., Jr Nat Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 9.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 10.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 11.Goodier JL, Ostertag EM, Kazazian HH., Jr Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 12.Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 14.Szak ST, Pickeral OK, Landsman D, Boeke JD. Genome Biol. 2003;4:R30. doi: 10.1186/gb-2003-4-5-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ejima Y, Yang L. Hum Mol Genet. 2003;12:1321–1328. doi: 10.1093/hmg/ddg138. [DOI] [PubMed] [Google Scholar]
- 16.Rozmahel R, Heng HH, Duncan AM, Shi XM, Rommens JM, Tsui LC. Genomics. 1997;45:554–561. doi: 10.1006/geno.1997.4968. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 18.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr Am J Hum Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chimpanzee Sequencing and Alnalysis Consortium. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 20.Boissinot S, Entezam A, Furano AV. Mol Biol Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- 21.Toomey RE, Wakil SJ. J Biol Chem. 1966;241:1159–1165. [PubMed] [Google Scholar]
- 22.Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- 23.Yang Z. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 24.Prince VE, Pickett FB. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 25.Dunn CA, Medstrand P, Mager DL. Proc Natl Acad Sci USA. 2003;100:12841–12846. doi: 10.1073/pnas.2134464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunn CA, van de Lagemaat LN, Baillie GJ, Mager DL. Gene. 2005;364:2–12. doi: 10.1016/j.gene.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 27.Esnault C, Maestre J, Heidmann T. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 28.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 30.Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 31.Krull M, Brosius J, Schmitz J. Mol Biol Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 32.Nekrutenko A, Li WH. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- 33.Cordaux R, Udit S, Batzer MA, Feschotte C. Proc Natl Acad Sci USA. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) Sunderland, MA: Sinauer; 2003. Version 4.0b10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.