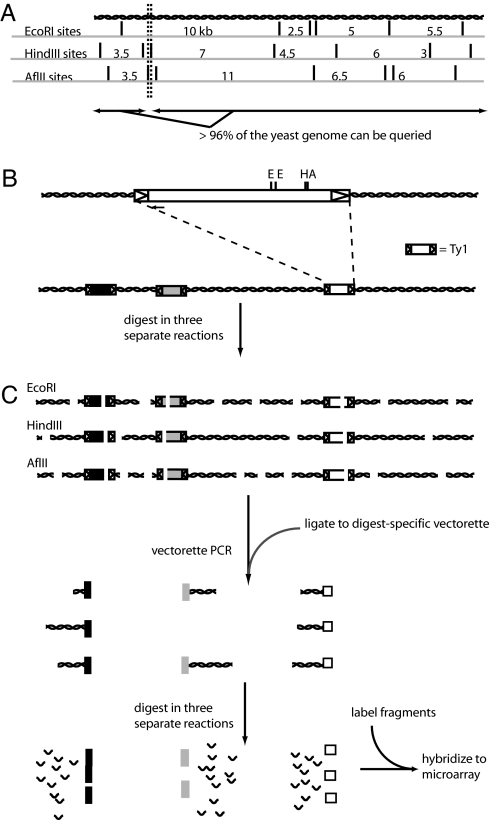
Fig. 1.
TIP-chip workflow. (A) Choosing restriction enzyme combinations for parallel digests of the yeast genome. The enzymes cut the DNA into overlapping pieces, so that each nucleotide of the yeast genome is contained in three separate restriction fragments, one from each enzyme. More than 96% of the yeast genome is contained in at least one fragment >1 kb and <10 kb; these somewhat arbitrary limits were chosen based on previous experience with PCR amplification and the proposed array design. (B) The Ty1 element, with LTRs shown as arrowheads. The small arrow at the 5′ end of the Ty1 denotes the position of the Ty1-specific primer used (JB8784; see supporting information). (C) Preparation of genomic DNA for hybridization to the TIP-chip. Genomic DNA is digested in three parallel reactions, with three restriction enzymes with 6-base recognition sequences. The digested fragments are ligated to digest-specific vectorettes and amplified by using vectorette PCR. Longer amplicons may not amplify well and may be underrepresented in the resulting mixture. The amplicons are then pooled and digested in three parallel reactions with three enzymes with 4-base recognition sequences. The resulting fragments are heat-inactivated, labeled, and hybridized to the microarray.