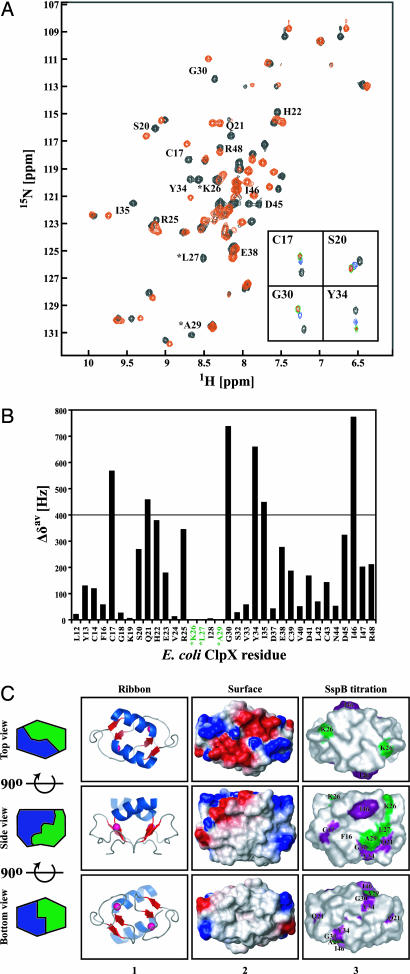
Fig. 2.
Binding of SspB154–165 to ZBD2 as monitored by NMR. (A) A heteronuclear sequential quantum correlation spectrum of 0.25 mM 15N-labeled ZBD2 in the absence (black) or presence of 5 mM of SspB154–165 (red). ∗, chemical shifts that disappear upon addition of peptide. Insets show the NH chemical shift changes of four residues in ZBD2 in the presence of 0 (black), 0.5 mM (blue), 2.5 mM (green), and 5 mM (red) of SspB154–165. (B) Shown are the chemical shift changes Δδav = [(Δδ1H)2 + (Δδ15N)2]1/2 in the presence of 5 mM SspB154–165. ∗, residues whose chemical shifts disappeared upon addition of 5 mM SspB154–165. (C) Ribbon and surface representation of ZBD dimer (Protein Data Bank ID code 1OVX) (5). Middle and Bottom are rotated 90o along the horizontal axis with respect to Top and Middle, respectively. In column 1, helices are colored blue, strands are red, and Zn(II) atoms are shown as pink spheres. In column 2, the electrostatic potential surface of ZBD2 is shown with negatively charged, positively charged, and hydrophobic surfaces in red, blue, and gray, respectively. In column 3, residues for which Δδav > 400 Hz and whose chemical shifts disappeared in the SspB154–165 titration experiments are colored purple and green, respectively. All structures were drawn by using PyMOL (http://pymol.sourceforge.net).