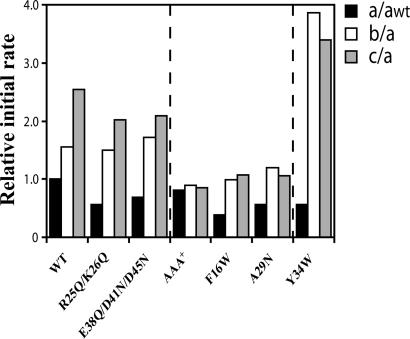
Fig. 3.
Mutational analysis to determine the SspB2-binding site on ZBD in ClpX. Relative initial rates of the ClpP-dependent degradation of GFP-SsrA (3.9 μM monomer concentration) mediated by ClpX or different ClpX mutants (each at 1 μM monomer concentration) in the presence of ClpP (1.2 μM monomer concentration) are shown in the presence of 0 (a), 0.025 μM (b), and 0.165 μM (c) of SspB2. The initial rates a were normalized to awt (black bars), whereas the initial rates b (white bars) and c (gray bars) were normalized to the a of the respective mutant. Initial rates were determined from the first 200 seconds of the curves shown in Fig. 10.