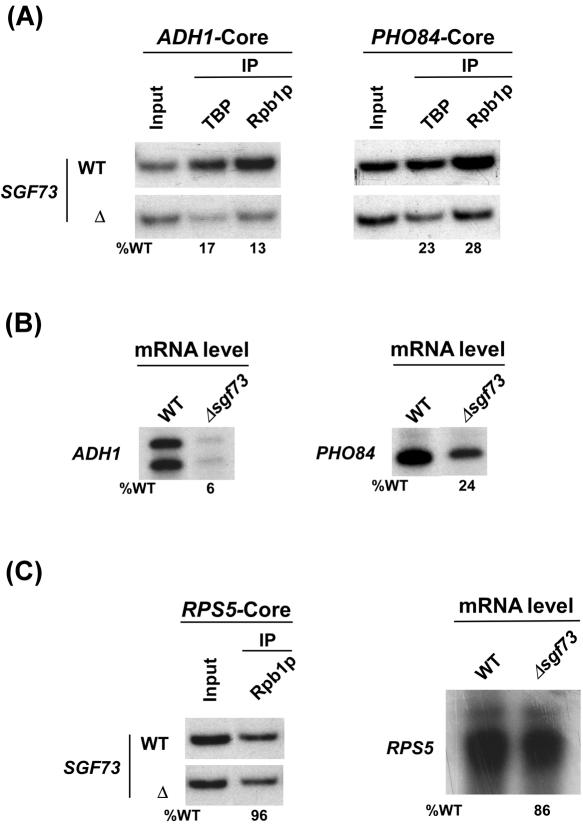
Figure 3.
Sgf73p is required for PIC formation (and hence transcriptional activation) at the ADH1 and PHO84 promoters. (A) The analysis of the PIC assembly at the ADH1 and PHO84 core promoters in Δsgf73. The yeast strains were grown in YPD at 30°C up to an OD600 of 1.0 prior to crosslinking. IPs were performed as described in Figure 2B. Primer-pairs located at the ADH1 and PHO84 core promoters were used for PCR analysis of the immunoprecipitated DNA samples. (B) Transcription. Total cellular RNA was prepared from the wild-type or Δsgf73, and mRNA levels from the ADH1 and PHO84 genes were quantitated by primer-extension. (C) Sgf73p is not required for PIC formation (and hence transcription) at the RPS5 promoter. Left panel: Analysis of recruitment of RNA polymerase II at the RPS5 core promoter in the wild-type and SGF73 deletion mutant strains. Both the wild-type and deletion mutant strains were grown, crosslinked and immunoprecipitated as in panel A. Primer-pair located at the RPS5 core promoter was used for PCR analysis of the IP DNA samples. Right panel: Transcription. Total cellular RNA was prepared from the wild-type or Δsgf73, and mRNA level from the RPS5 gene was quantitated by primer-extension.