Figure 1.
A Genetic Screen for Functional Gene Transfer from the Plastid Genome to the Nucleus.
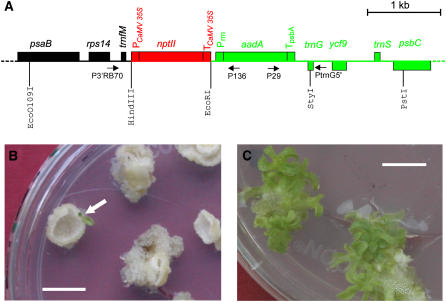
(A) Physical map of the region in the plastid genome transferred to the nuclear genome by selection for kanamycin resistance (Stegemann et al., 2003). The chloroplast-type spectinomycin/streptomycin resistance gene aadA and flanking chloroplast sequences are shown in green. The eukaryotic-type nptII gene mimics an upstream nuclear gene (red) in that it simulates the landing of the chloroplast aadA gene downstream of a resident nuclear gene. Genes above the line are transcribed from the left to the right, and genes below the line are transcribed in the opposite orientation. Relevant restriction sites and primers are also shown, and the orientation of PCR primers is indicated by arrows. PCaMV 35S, cauliflower mosaic virus 35S promoter; TCaMV 35S, cauliflower mosaic virus 35S terminator; Prrn, chloroplast rRNA operon promoter; TpsbA, terminator from the chloroplast psbA gene.
(B) Selection for functional activation of the transferred aadA gene. Primary lines with an activated aadA gene (arrow) were identified by large-scale selection experiments on plant regeneration medium containing both spectinomycin and streptomycin. Resistance to these antibiotics is dependent on the aadA gene product, an enzyme inactivating aminoglycoside antibiotics.
(C) Shoot regeneration from tissue pieces of the primary line in the presence of 500 μg/mL spectinomycin indicates high-level antibiotic resistance. Bars = 1 cm.