Figure 1.
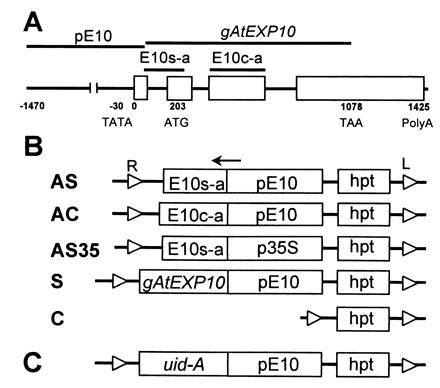
Structure of AtEXP10 and antisense, sense, and control constructs for Arabidopsis transformation. (A) Gene structure of AtEXP10. Open boxes represent exons. E10s-a and E10c-a indicate the regions from which antisense sequences were prepared for AS (or AS35) and AC, respectively. A sense sequence was prepared from gAtEXP10. pE10 represents the promoter region of AtEXP10. (B) Transgene constructs. AS, antisense construct specific to AtEXP10 (E10s-a) driven by pE10; AC, antisense construct containing AtEXP10 coding region (E10c-a) and driven by pE10; AS35, antisense construct specific to AtEXP10 (E10s-a) driven by 35S CaMV promoter (p35S); S, sense construct containing genomic AtEXP10 (gAtEXP10) and driven by pE10; C, control construct containing only hygromycin phosphotransferase gene cassette (hpt) as a selectable marker. Small open triangles at the end of the each line represent right border (R) and left border (L) of the T-DNA in Agrobacterium Ti (tumor-inducing) plasmid. The arrow indicates the direction of transcription. (C) The construct for expression of GUS driven by pE10 (AtEXP10∷GUS).
