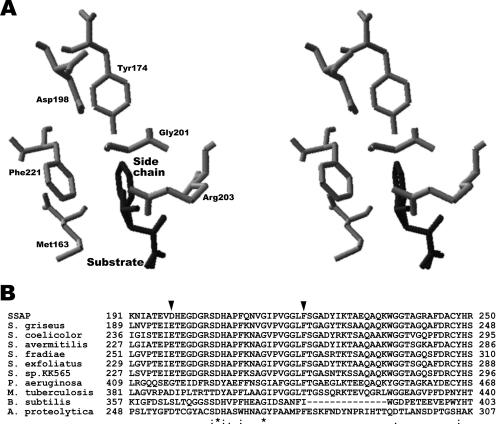
FIG. 1.
Predicted local three-dimensional structure of SSAP around bound substrates and alignment of SSAP sequence around Asp198 and Phe221 with the corresponding regions of other bacterial aminopeptidases. (A) All residues and substrates are shown using stick models. The residues surrounding the side chain of the substrate are shown in gray and the bound substrates (free Leu and Phe) in dark gray. (B) The two residues mutated in this study are indicated by black arrowheads. *, conserved residues; :, conservative substitutions; ., semiconservative substitutions in all sequences. Proteins: S. griseus, aminopeptidase from Streptomyces griseus; S. coelicolor, putative aminopeptidase from Streptomyces coelicolor; S. avermitilis, putative aminopeptidase from Streptomyces avermitilis; S. fradiae, aminopeptidase from Streptomyces fradiae; S. exfoliatus, aminopeptidase from Streptomyces exfoliatus; S. sp.KK565, aminopeptidase from Streptomyces sp. strain KK565; P. aeruginosa, a secreted aminopeptidase from Pseudomonas aeruginosa; M. tuberculosis, aminopeptidase from Mycobacterium tuberculosis; B. subtilis, extracellular aminopeptidase from Bacillus subtilis; A. proteolytica, leucine aminopeptidase from Aeromonas proteolytica. Multiple sequence alignments were performed with the CLUSTAL algorithm (http://www.ddbj.nig.ac.jp/search/clustalw-j.html).