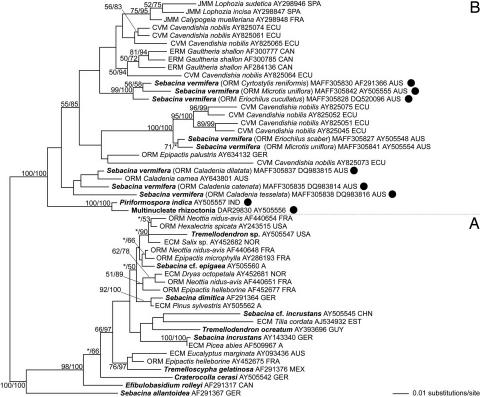
Fig. 1.
Phylogenetic placement of the strains tested in this study within the Sebacinales, estimated by maximum likelihood from an alignment of nuclear rDNA coding for the 5′ terminal domain of the ribosomal large subunit. Branch support is given by nonparametric maximum likelihood bootstrap (first numbers) and by posterior probabilities estimated by Bayesian Markov chain Monte Carlo (second numbers). Support values of <50% are omitted or indicated by an asterisk. The tree was rooted according to the results of ref. 11, and subgroups discussed in ref. 11 are denoted with “A” and “B.” Sequences of the strains used in this study are indicated by black circles. Sequences from morphologically determined specimens or cultures are printed in bold. Sebacinalean sequences obtained from mycorrhizal plant roots are assigned to mycorrhizal types by the following acronyms: CVM, cavendishioid mycorrhiza (14); ECM, ectomycorrhiza; ERM, ericoid mycorrhiza; JMM, jungermannioid mycorrhiza; and ORM, orchid mycorrhiza. Proveniences are given as follows: A, Austria; AUS, Australia; CAN, Canada; CHN, People's Republic of China; ECU, Ecuador; EST, Estonia; FRA, France; GER, Germany; GUY, Guyana; IND, India; MEX, Mexico; NOR, Norway; and SPA, Spain.