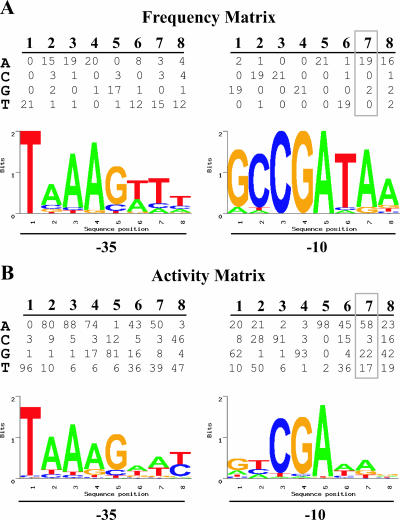
FIG. 2.
Probability weight matrices and sequence logos for predicting promoters in the chlamydial genome. (A) Frequency matrix for the 16 positions in the −35 and −10 promoter elements and the four possible nucleotides at each position (see Materials and Methods). Each value in the matrix indicates how many of the 21 known σ28 promoters in the training set contained the given nucleotide at that promoter position. A sequence logo depicting the relative nucleotide preference at each position in the promoter is shown below the matrix (25). (B) Activity matrix for the −35 and −10 promoter elements with values derived from a mutational analysis of the C. trachomatis hctB promoter as described in the text (25). At each promoter position, the values are proportional to the relative promoter activity attributable to that nucleotide for a total of 100%. The sequence logo for the promoter is shown below the matrix. Details of the sequence logo format are presented in Materials and Methods and Results. All sequence logos were derived using SEQLOGO, which is available online at http://ep.ebi.ac.uk/EP/SEQLOGO/.