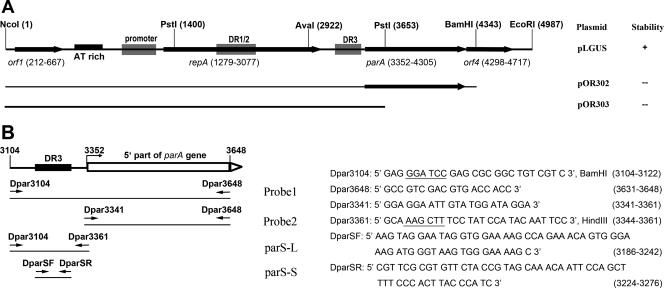
FIG. 1.
(A) Physical map of the pCXC100 replicon and the composition of the pCXC100 replicon in the pBR325-based plasmids used in this study. Plasmid pLGUS contains the complete replicon, while pOR302 contains a deleted replicon lacking the majority of orf4 and pOR303 lacks both the main body of parA and the whole orf4. Other data shown in this study indicate that orf4 encodes a ParB analogue that has specific DNA-binding activity. Whether each plasmid stably propagated in L. xyli subsp. cynodontis is listed on the right: +, the plasmid was stable when propagating in the bacterium; −, the plasmid was not stable. The details of the plasmid stability are shown in Table 1. (B) Probes used in this study to assess the DNA-binding activity encoded by orf4. The positions and sequences of primers used for PCR amplification of the corresponding DNA fragments are indicated. Restriction sites for Dpar3104 and Dpar3361, given after the sequences, are underlined. DR, direct repeat.