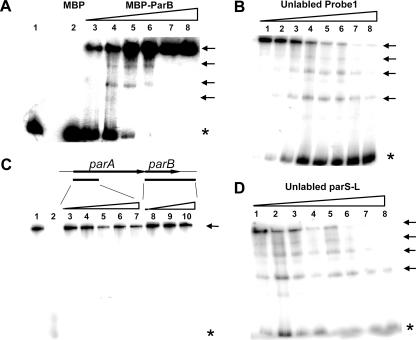
FIG. 2.
Electrophoretic mobility shift assay of the specific DNA-binding activity of the ParB protein in vitro. All DNA probes used here were PCR amplified and radiolabeled at the 5′ end with [γ-32P]ATP. Probes are defined in Fig. 1B. (A) Dose-dependent binding of the ParB protein with probe 1. Increasing amounts of MBP-ParB fusion protein (from lane 3 to lane 8, 100, 200, 400, 800, 1,600, and 1,900 ng, respectively) were incubated with a fixed amount of radiolabeled probe 1 DNA fragment. Control samples in lanes 1 and 2 contained 1,000 ng of BSA protein and MBP, respectively. (B) Assay of the competition ability of the unlabeled DNA probe 1 with the labeled probe 1. The ratios of the radiolabeled to unlabeled DNA for the reactions were (from lanes 1 to 8, respectively) 1:0, 1:1, 1:2.5, 1:5, 1:7.5, 1:10, 1:15, and 1:27. (C) Assay of the ability of the unlabeled probe 2 (5′ part of parA) and parB (containing the full parB gene and 280 bp of the downstream sequence) to compete with the labeled probe 1. The ratios of labeled probe 1 to unlabeled probe 2 (from lanes 3 to 7, respectively) were 1:1, 1:5, 1:10, 1:19, and 1:37, and those to the unlabeled parB gene (from lanes 8 to 10, respectively) were 1:1, 1:5, and 1:19. Lane 1 contained no unlabeled DNA. The reaction shown in lane 2 was conducted in the same manner as that shown in lane 8 of panel B. (D) Assay of the competition ability of the unlabeled DNA parS-L with the labeled probe 1. The ratios of the radiolabeled to unlabeled DNA were the same as those given for panel B. The amount of MBP-ParB fusion protein for each reaction shown in panels B to D was 200 ng. Asterisks indicate unbound DNA, and arrows indicate different forms of protein-DNA complexes.