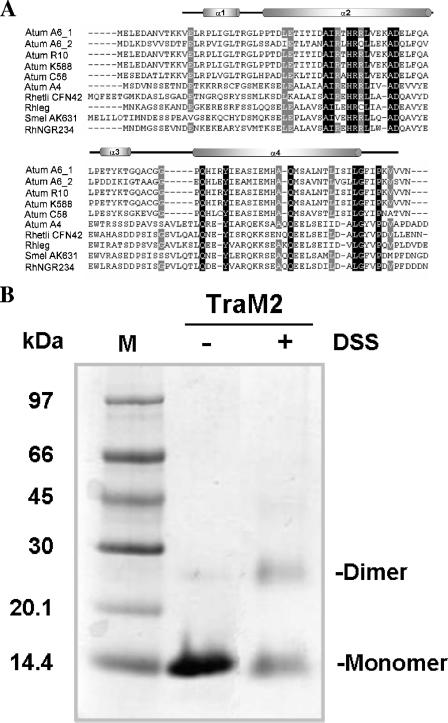
FIG. 1.
Sequence alignment and structural analysis of TraM2. (A) Sequence alignment of TraM proteins using GeneDoc (16). Amino acid sequences are from the following bacteria: (i) Atum A6_1, A. tumefaciens A6; (ii) Atum A6_2, A. tumefaciens A6, traM2; (iii) Atum R10, A. tumefaciens R10; (iv) Atum K588, A. tumefaciens K588; (v) Atum C58, A. tumefaciens C58; (vi) Atum A4, A. tumefaciens A4; (vii) Rhetli CFN42, Rhizobium etli CFN42; (viii) Rhleg, Rhizobium leguminosarum; (ix) Smel AK631, Sinorhizobium meliloti AK631; and (x) RhNGR234, Rhizobium sp. NGR234. Invariant residues are highlighted in black, and highly conserved resides are shaded in gray. Secondary structure elements of TraM2 are indicated above. (B) Cross-linking of TraM2 with DSS. TraM2 at a final concentration of 2.86 mg/ml was incubated in the presence (+) or absence (−) of 25 mM DSS for 30 min at room temperature and quenched with 1 M Tris-Cl, pH 8.0, and then subjected to 15% Tris-Tricine gel electrophoresis. The protein bands were visualized by staining with Coomassie brilliant blue. The bands corresponding to monomer and dimer molecules are indicated. M, molecular weight marker.