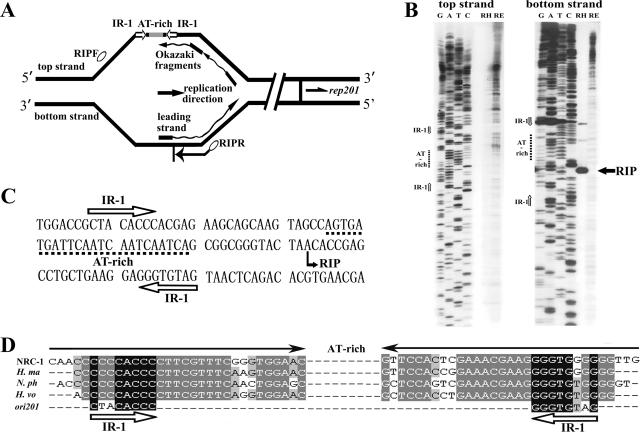
FIG. 5.
Mapping of the RIP of pSCM201. (A) Outline of RIP mapping for the unidirectional theta replication of pSCM201 (not to scale). Nascent DNAs (leading strand and Okazaki fragments) initiated by a small RNA primer (black rectangles) were used as the templates in the primer extension reaction to determine the RIP. Ovals indicate the primers used in RIP mapping. The directions of pSCM201 replication, rep201 transcription, and primer extension are indicated by arrows. (B) Identification of the replication initiation point by RIP mapping. Primer extension was performed with the oligonucleotides RIPF for the top strand (left panel) and RIPR for the bottom strand (right panel). Lanes contain dideoxy sequencing reaction mixtures (GATC) and the products of the primer extension of pSCM201 RIs from Haloarcula sp. strain AS7094 (RH) and pSCM2011 RIs from E. coli (RE). The filled arrow indicates the RIP position of pSCM201. The open arrows indicate the positions of IR-1. The dashed lines indicate the positions of the AT-rich region. (C) Nucleotide sequence of a region containing IR-1, the AT-rich region, and the RIP of pSCM201. (D) Conserved inverted repeats in ori201 and the chromosomal replication origins of the four other haloarchaea. The filled arrows indicate the long inverted repeat flanking the AT-rich region in the four haloarchaeal chromosomal replication origins. The open arrows indicate IR-1 of ori201, which shares high homology with the inner sequence of those chromosomal long inverted repeats. NRC-1, Halobacterium sp. strain NRC-1; H. ma, Haloarcula marismotui; N. ph, Natronomonas pharaonis; H. vo, Haloferax volcanii.