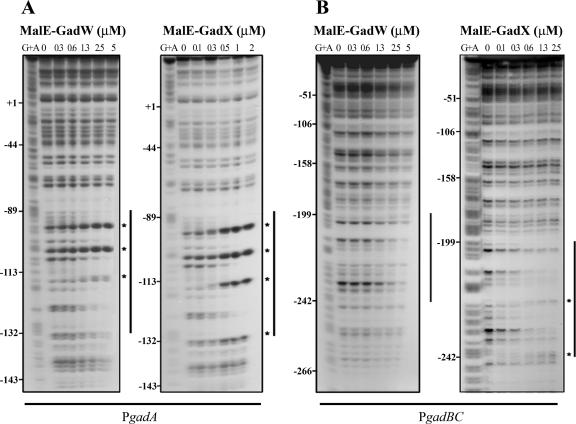
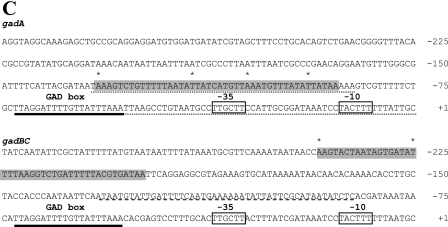
FIG.4.
Identification of MalE-GadW and MalE-GadX binding sites in gadA and gadBC regulatory regions. DNase I footprinting assays were performed using PgadA (positions −176 to 77) (A) and PgadBC (positions −307 to 77) (B) with different amounts of MalE-GadW and MalE-GadX. Samples were processed as described in Materials and Methods. The protected regions are indicated by vertical lines, and the hypersensitive sites are indicated by asterisks. (C) Nucleotide sequences of the gadA and gadBC regulatory regions. The MalE-GadX and MalE-GadW binding sites, identified by DNase I footprinting assays (panels A and B), are indicated by shading, and the hypersensitive sites are indicated by asterisks. The low-affinity MalE-GadX site in the gadBC promoter (37) is underlined with a dashed line. The H-NS binding sites in the gadA promoter (14) are underlined with dotted lines. The −35 and −10 hexamers are enclosed in boxes. The GAD box is underlined with a boldface line in both promoter regions.