Abstract
The rate-limiting enzyme of anaerobic benzoate degradation by Rhodopseudomonas palustris, benzoyl coenzyme A (CoA) reductase, is highly sensitive to oxygen, and its synthesis is tightly regulated. We determined that a previously unknown gene in the benzoate gene cluster, badM, encodes a transcriptional repressor of benzoyl-CoA reductase gene expression. BadM controls gene expression from the benzoyl-CoA reductase promoter in concert with two previously described transcriptional activators.
Under anaerobic conditions some bacteria catabolize structurally diverse aromatic compounds through peripheral pathways to form benzoate or benzoyl coenzyme A (CoA), which then enters a central pathway of aromatic ring reduction and ring cleavage (12). A novel oxygen-sensitive enzyme, benzoyl-CoA reductase, is critical for the operation of the pathway, because it relieves the resonance stability of the benzene ring (5, 11). The photosynthetic bacterium Rhodopseudomonas palustris is one of several species that have served as model organisms for studies of anaerobic benzoate degradation (2, 15). The R. palustris benzoate degradation gene cluster is unique in that it encodes a small protein, named BadM, which belongs to the Rrf2 family of transcriptional regulators (PFAM: PF02082 and Interpro: IPR000944) (16) (Fig. 1). Several Rrf2 family members have been shown to function as repressors, and we present evidence here that BadM (RPA0663) represses the transcription of benzoate degradation genes. A badM mutation resulted in constitutive expression of the badDEFGAB operon, encoding benzoyl-CoA reductase and benzoate-CoA ligase. We had previously shown that transcription of the benzoyl-CoA reductase operon is activated by the regulators AadR and BadR in response to anaerobiosis and benzoate, respectively (6, 9). BadM is thus a third regulator that acts at the badDEFGAB promoter.
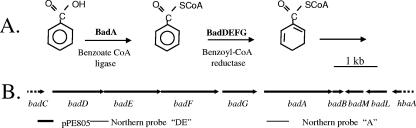
FIG. 1.
(A) The first two steps of anaerobic benzoate degradation in R. palustris. (B) The organization of some of the benzoate degradation genes in R. palustris. Arrows indicate the direction of transcription. The DNA fragment used to construct the promoter fusion plasmid pPE805 is indicated, as are the fragments used as probes for the Northern hybridizations.
A badM mutant expresses the first two enzymes of anaerobic benzoate degradation constitutively.
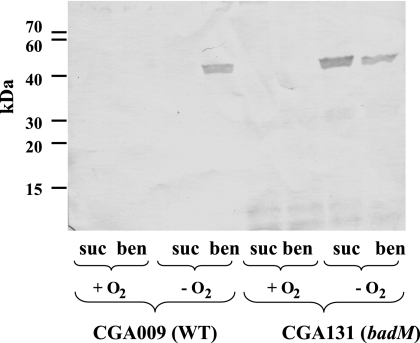
To examine the possible contribution of BadM to benzoate degradation, we used standard techniques (14) to construct a mutant (CGA131) in which the badM gene had been deleted from the chromosome (Table 1). Southern hybridization experiments verified that the expected deletion had occurred. The badM deletion mutant grew as well as its wild-type parent in minimal medium with 3 mM benzoate under anaerobic conditions in light, and it also grew normally in minimal medium with 10 mM succinate both aerobically and anaerobically in light (17). The enzymes of anaerobic benzoate degradation are induced when cells are grown anaerobically on benzoate and are not present at significant levels in succinate-grown cells (9, 22). Rabbit polyclonal antisera were raised against purified polyhistidine-tagged subunits of benzoyl-CoA reductase (His-BadD and His-BadG) at Covance Research Products Inc. (Denver, PA) and used to screen for a badM mutant phenotype. Cell extracts (20 μg protein) of R. palustris were separated on 12% sodium dodecyl sulfate-polyacrylamide gels and immunoblotted as described previously (22). Immunoblot analysis indicated that the badM mutant differed from the wild type in that it synthesized the BadD subunit of benzoyl-CoA reductase when it was grown anaerobically with succinate as well as when it was grown with benzoate (Fig. 2). Immunoblots probed with antisera against either the BadG subunit of benzoyl-CoA reductase or benzoate-CoA ligase (BadA) (17) showed a similar pattern of constitutive protein production in the badM mutant (data not shown). In all cases anaerobic growth was prerequisite for constitutive enzyme production. The badM mutation did not cause constitutive production of three enzymes for anaerobic benzoate degradation that function downstream of the benzoyl-CoA reductase step (e.g., BadK, cyclohex-1-ene-1-carboxyl-CoA hydratase; BadH, 2-hydroxycyclohexane carboxyl-CoA dehydrogenase; and BadI, 2-ketocyclohexane carboxyl-CoA hydrolase) (15) as determined by Western blot analysis (data not shown). Constitutive expression of benzoate-CoA ligase activity was observed in succinate-grown cells of the badM mutant (Table 2), confirming the results from immunoblot analyses. When BadM was expressed in trans in the badM mutant, repression of benzoate-CoA ligase activity in succinate-grown cells was restored. Immunoblot analysis indicated that BadM expressed in trans also complemented the constitutive expression phenotype of benzoyl-CoA reductase (data not shown).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant propertya | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | Host strain for cloning | Gibco-BRL |
| S17-1 | Donor strain for transferring broad-host-range plasmids | 26 |
| P. aeruginosa PAO1 | Wild-type strain | A. Kropinsky |
| R. palustris | ||
| CGA009 | Wild-type strain; spontaneous Cmr derivative | 19 |
| CGA131 | badM (257-bp deletion mutant) | This study |
| CGA901 | badA::′lacZ; Kmr | This study |
| CGA301 | badM badA::′lacZ; Kmr | This study |
| Plasmids | ||
| pBBR1MCS-2 | Broad-host-range vector; Kmr | 18 |
| pBBR1MCS-5 | Broad-host-range vector; Gmr | 18 |
| pJQ200mp18 | Mobilizable suicide vector; sacB Gmr | 23 |
| pHRP309 | Reporter plasmid, contains promoterless ′lacZ | 21 |
| pCP230 | pJQ200mp18 with badA::′lacZ; Kmr | This study |
| pCP439 | pBBR1MCS-2 with the badM gene inserted; Kmr | This study |
| pCP561 | pBBR1MCS-5 with the badM gene inserted; Gmr | This study |
| pPE805 | pHRP309 with badD promoter region fused to ′lacZ; Spr Gmr | This study |
Gmr, gentamicin resistance; Kmr, kanamycin resistance; Cmr, chloramphenicol resistance; Spr, spectinomycin resistance. Antibiotic concentrations were as described previously (24).
FIG. 2.
Western immunoblot of cell extracts of R. palustris wild type (WT; CGA009) and a badM mutant (CGA131) grown under different conditions and probed with BadD (first subunit of benzoyl-CoA reductase) antiserum. + O2, grown in aerobic conditions; − O2, grown in anaerobic conditions. Cells were grown with 10 mM succinate (suc) or with 10 mM succinate plus 3 mM benzoate (ben). Each lane contains 20 μg protein. Size markers are indicated on the left in kilodaltons.
TABLE 2.
Effect of badM on benzoate-CoA ligase activity
| Strainb | Benzoate-CoA ligase activity for cellsa
|
|
|---|---|---|
| Benzoate grown | Succinate grown | |
| CGA009 | 30.2 | 6.0 |
| CGA131 | 27.8 | 55.9 |
| CGA009(pCP561) | 27.7 | 7.5 |
| CGA131(pCP561) | 28.7 | 8.5 |
Expressed as nmol product per min per mg protein in crude cell extracts (8). The concentration of benzoate added to the reaction mixtures was 10 μM. Values are the averages of activities from two independently prepared extracts.
CGA009, wild type; CGA131, badM mutant.
BadM represses at the level of transcription.
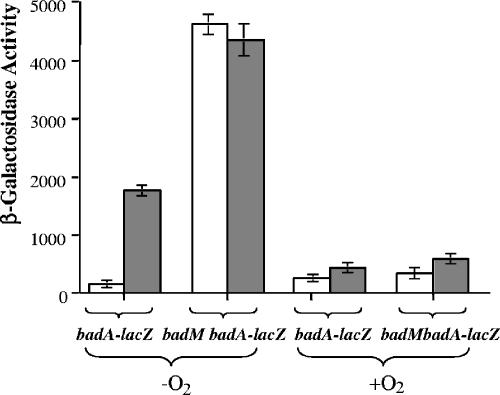
Total RNA was isolated from R. palustris cells using the High Pure RNA isolation kit (Roche Diagnostics Corp., Indianapolis, IN), and 10 μg RNA was used per lane for Northern blot analysis. The analysis was performed with the NorthernMax-Gly kit (Ambion, Austin, TX) according to the manufacturer's instructions. A 440-bp PCR product containing the junction of badD and badE and a 522-bp PCR product targeting badA (Fig. 1) were labeled with [α-32P]dCTP using Ready-To-Go DNA labeling beads (Amersham, Arlington Heights, IL), purified with a ProbeQuant G-50 Microcolumn (Pharmacia Biotech, Piscataway, NJ), and used as probes. A single transcript of about 7,000 bp was present at much higher levels in succinate-grown badM cells than in wild-type cells (data not shown). The predicted size of a badDEFGAB transcript is 6,758 bp (19). BadB encodes a ferredoxin that likely functions with benzoyl-CoA reductase. This gene is separated from badA by just 16 bp. Together our results indicate that BadM acts at the level of transcription to repress expression of a badDEFGAB operon. Consistent with this we found that in a wild-type background benzoate induced the expression of a badA::lacZ chromosomal transcriptional fusion 12-fold above the level seen in succinate-grown cells (Fig. 3). When the badM gene was mutated, there was no significant difference in lacZ expression levels between induced and noninduced cells and the levels of β-galactosidase were much higher than the noninduced wild-type level. Significant levels of badA expression were seen only under anaerobic conditions.
FIG. 3.
Expression of β-galactosidase activity from a badA::′lacZ chromosomal fusion in R. palustris wild-type and badM mutant cells. Cells were grown with succinate (open bars) or on succinate plus benzoate (filled bars). Data are averages from three different experiments, plus or minus standard deviations. β-Galactosidase activity is shown as specific activity (nanomoles o-nitrophenol formed per min per mg protein) (9).
BadM represses transcription from the badD promoter.
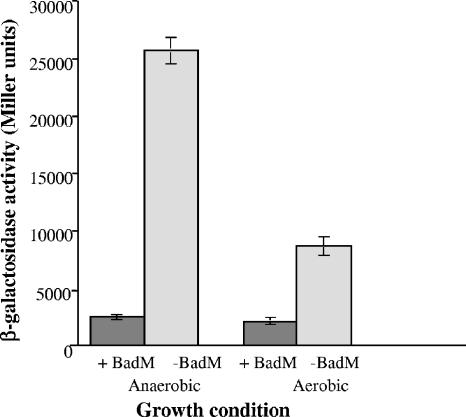
We constructed a badD promoter-lacZ transcriptional fusion plasmid according to methods described previously (10) and introduced it into Pseudomonas aeruginosa together with a plasmid expressing badM to test whether BadM was sufficient to regulate gene expression in a heterologous background. BadM repressed PbadD-lacZ expression in P. aeruginosa cells grown anaerobically in Luria broth under denitrifying conditions (Fig. 4). Addition of benzoate to the growth medium did not cause an increase in PbadD-lacZ expression. The repressive effect of BadM was also seen but was much less pronounced in aerobically grown cells. Previously we have shown that the badD promoter is regulated by an Anr/Fnr homologue called AadR in response to anaerobiosis (9). It is possible that the transcription factor Anr activates PbadD expression in anaerobically grown P. aeruginosa cells in the absence of BadM.
FIG. 4.
Expression of β-galactosidase activity in P. aeruginosa cells containing the PbadD-′lacZ reporter plasmid pPE805 and either the BadM-expressing plasmid pCP439 (+ BadM) or the control vector pBBR1MCS-2 (−BadM). Cells were grown aerobically in Luria broth or anaerobically in Luria broth plus 0.2% glucose plus 10 mM potassium nitrate. Data are averages from three different experiments, plus or minus standard deviations. β-Galactosidase activity is in Miller units (20).
Possible mechanism of BadM activity.
Our description of the BadM protein expands the range of functions regulated by Rrf2 proteins to include anaerobic biodegradation. Other characterized Rrf2 family regulators are repressors of genes involved in nitrite, nitric oxide, or iron metabolism (3, 4, 13, 25, 27). The best-studied Rrf2 protein, IscR, contains a Fe-S cluster and represses the expression of several Fe-S-cluster-containing anaerobic respiration enzymes in addition to regulating Fe-S cluster formation genes (13, 25). Three cysteine residues that are conserved in the C-terminal portion of IscR and other Rrf2 regulators have been proposed to be involved in the formation of its Fe-S cluster (4, 25). Although BadM has the Rrf2-type helix-turn-helix domain signature presumed to be involved in DNA binding, it does not have the conserved cysteines that are found in many members of this family. In a transcriptome analysis using R. palustris Affymetrix gene chips we found that the expression of only a few genes in addition to the benzoyl-CoA reductase operon (expressed at 20-fold-higher levels in a badM mutant grown anaerobically on succinate than in wild-type cells grown the same way) was affected by more than fivefold by badM. These included a dicarboxylic acid transporter gene (RPA2448) and a gene for a conserved hypothetical protein (RPA3401) that were expressed at about sevenfold- and sixfold-higher levels in a badM mutant, respectively. badM was required for full expression of a possible cytochrome P450 gene (RPA1009; eightfold effect) as well as an operon of conserved hypothetical genes (RPA1209-1212; sixfold effect). None of these genes appear to encode Fe-S proteins or to be involved in iron-related metabolism.
We were surprised to find that BadM had such a large effect in repressing benzoyl-CoA reductase expression because we had previously described two regulators, AadR (which senses anaerobiosis) and BadR (which senses benzoate or benzoyl-CoA), that appeared to account for the full range of benzoate-induced expression of the badDEFG genes (9). A badR aadR double mutant is completely defective in growth on benzoate, indicating an absolute requirement for these regulators (9). The anaerobic benzoate degradation genes of Azoarcus sp. strain CIB, a denitrifying bacterium, are induced by AcpR, a regulator that is related to AadR, in response to oxygen (7), and by BzdR, a transcriptional repressor that is not related to BadM (1), in response to benzoyl-CoA. It is logical to hypothesize that BadM represses benzoate degradation by binding to the badD promoter and that benzoate binds to BadM to cause it to come off DNA and derepress gene expression in R. palustris, but we have not been able to show this. The addition of benzoate to P. aeruginosa cells harboring BadM and a badD-lacZ promoter fusion did not have an effect on lacZ expression. This could be because the effector of BadM is a compound (such as benzoyl-CoA) other than benzoate, or it could be that BadM interacts with BadR or AadR or their DNA binding sites in a more complex way to modulate gene expression. It will be important to work with purified regulators in vitro to determine their DNA binding sites and define possible physical interactions between regulatory proteins and RNA polymerase in order to fully understand the mechanism of oxygen- and benzoate-mediated regulation at the badDEFGAB promoter.
Acknowledgments
This work was supported by the Division of Energy Biosciences, U.S. Department of Energy (grant DE-FG02-05ER15707), and by the U.S. Army Research Office (grant W911NF-05-1-0176). C.M.P. thanks the Belgian American Educational Foundation (BAEF), the D. Collen Research Foundation, and the Firmin van Brée of the Hoover Foundation for the University of Louvain for a postdoctoral fellowship.
We thank Janelle Torres y Torres for constructing and purifying His-BadD and His-BadG.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Barragan, M. J., B. Blazquez, M. T. Zamarro, J. M. Mancheno, J. L. Garcia, E. Diaz, and M. Carmona. 2005. BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 280:10683-10694. [DOI] [PubMed] [Google Scholar]
- 2.Barragán, M. J. L., M. Carmona, M. T. Zamarro, B. Thiele, M. Boll, G. Fuchs, J. L. García, and E. Díaz. 2004. The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J. Bacteriol. 186:5762-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaumont, H. J., S. I. Lens, W. N. Reijnders, H. V. Westerhoff, and R. J. van Spanning. 2004. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol. Microbiol. 54:148-158. [DOI] [PubMed] [Google Scholar]
- 4.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188:874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boll, M., and G. Fuchs. 1995. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur. J. Biochem. 234:921-933. [DOI] [PubMed] [Google Scholar]
- 6.Dispensa, M., C. T. Thomas, M.-K. Kim, J. A. Perrotta, J. Gibson, and C. S. Harwood. 1992. Anaerobic growth of Rhodopseudomonas palustris on 4-hydroxybenzoate is dependent on AadR, a member of the cyclic AMP receptor protein family of transcriptional regulators. J. Bacteriol. 174:5803-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durante-Rodriguez, G., M. T. Zamarro, J. L. Garcia, E. Diaz, and M. Carmona. 2006. Oxygen-dependent regulation of the central pathway for the anaerobic catabolism of aromatic compounds in Azoarcus sp. strain CIB. J. Bacteriol. 188:2343-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egland, P. G., J. Gibson, and C. S. Harwood. 1995. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J. Bacteriol. 177:6545-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egland, P. G., and C. S. Harwood. 1999. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an Fnr family member. J. Bacteriol. 181:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egland, P. G., and C. S. Harwood. 2000. HbaR, a 4-hydroxybenzoate sensor and FNR-CRP superfamily member, regulates anaerobic 4-hydroxybenzoate degradation by Rhodopseudomonas palustris. J. Bacteriol. 182:100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egland, P. G., D. A. Pelletier, M. Dispensa, J. Gibson, and C. S. Harwood. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. USA 94:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu. Rev. Microbiol. 56:345-369. [DOI] [PubMed] [Google Scholar]
- 13.Giel, J. L., D. Rodionov, M. Liu, F. R. Blattner, and P. J. Kiley. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 60:1058-1075. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, F. H., and C. S. Harwood. 2005. The pimFABCDE operon from Rhodopseudomonas palustris mediates dicarboxylic acid degradation and participates in anaerobic benzoate degradation. Microbiology 151:727-736. [DOI] [PubMed] [Google Scholar]
- 15.Harwood, C. S., G. Burchhardt, H. Herrmann, and G. Fuchs. 1999. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22:439-458. [Google Scholar]
- 16.Keon, R. G., R. Fu, and G. Voordouw. 1997. Deletion of two downstream genes alters expression of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough. Arch. Microbiol. 167:376-383. [DOI] [PubMed] [Google Scholar]
- 17.Kim, M.-K., and C. S. Harwood. 1991. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol. Lett. 83:199-204. [Google Scholar]
- 18.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, I. I., and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 19.Larimer, F. W., P. Chain, L. Hauser, J. Lamerdin, S. Malfatti, L. Do, M. L. Land, D. A. Pelletier, J. T. Beatty, A. S. Lang, F. R. Tabita, J. L. Gibson, T. E. Hanson, C. Bobst, J. L. Torres, C. Peres, F. H. Harrison, J. Gibson, and C. S. Harwood. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22:55-61. [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1992. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Parales, R. E., and C. S. Harwood. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram- bacteria. Gene 133:23-30. [DOI] [PubMed] [Google Scholar]
- 22.Pelletier, D. A., and C. S. Harwood. 1998. 2-Ketocyclohexanecarboxyl coenzyme A hydrolase, the ring cleavage enzyme required for anaerobic benzoate degradation by Rhodopseudomonas palustris. J. Bacteriol. 180:2330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 24.Rey, F. E., Y. Oda, and C. S. Harwood. 2006. Regulation of uptake hydrogenase and effects of hydrogen utilization on gene expression in Rhodopseudomonas palustris. J. Bacteriol. 188:6143-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98:14895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 27.Yeoman, K. H., A. R. Curson, J. D. Todd, G. Sawers, and A. W. Johnston. 2004. Evidence that the Rhizobium regulatory protein RirA binds to cis-acting iron-responsive operators (IROs) at promoters of some Fe-regulated genes. Microbiology 150:4065-4074. [DOI] [PubMed] [Google Scholar]