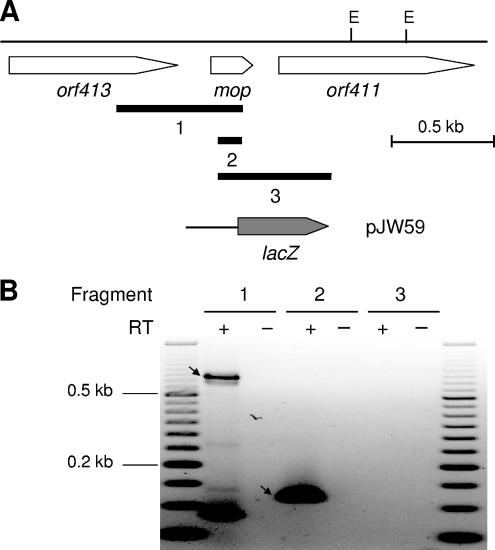
FIG. 3.
Transcriptional analysis of the R. capsulatus mop gene region. (A) Physical and genetic maps of the mop gene region. The physical map is given for EcoRI (E). Black bars below the genetic map indicate DNA fragments 1 to 3 emerging from RT-PCR (see Materials and Methods and panel B). The corresponding primer pairs used for RT-PCR are listed in Table 2. Hybrid plasmid pJW59, carrying a transcriptional mop-lacZ fusion, is based on the mobilizable broad-host-range plasmid pML5. (B) Transcriptional analysis of the mop gene region by RT-PCR. Total RNA was isolated from R. capsulatus cells grown in the presence of 10 μM Na2MoO4. Either RNA samples were treated with reverse transcriptase to synthesize cDNA (+) or, as a negative control, reverse transcriptase was omitted (−). Amplification products corresponding to DNA fragments 1 and 2 are marked by arrows. A 50-bp DNA ladder (Fermentas, St. Leon-Rot, Germany) was used as a length standard.