Abstract
A TaqMan real-time PCR assay, the COBAS TaqMan human immunodeficiency virus (HIV) (HPS-CTMHIV) PCR assay, recently developed for the quantification of HIV type 1 RNA in plasma, was evaluated in comparison with the licensed COBAS AMPLICOR HIV-1 MONITOR (CAHIM) assay. In this study, we have analyzed the tests' sensitivities, precisions, and linearities using multiple replicates of a standard panel of HIV RNA covering a 7-logarithm range of concentrations, as well as serial threefold dilutions of high-titer clinical samples. The subtype inclusivity was also investigated, using a panel of subtypes A to H, while a collection of 160 clinical samples was analyzed to assess the tests' specificities and the systems' similarities. The results of these experiments showed that the HPS-CTMHIV assay has a sensitivity of 53 copies/ml (95% hit rate), 100% specificity, and good intra- and interassay precision. The results of the HPS-CTMHIV assay were linear in the 50- to 107-copies/ml range, with a correlation coefficient (R) for expected versus observed results of 0.98. Compared to the CAHIM assay, the HPS-CTMHIV assay showed a high correlation (R = 0.99) across the dynamic range of RNA concentrations that, for the CAHIM assay, requires two different sample preparations. Equivalent performances were also observed for the two systems in the detection and quantification of HIV subtypes A to H. These data indicate that the HPS-CTMHIV assay may be one of the tests of choice for monitoring viral load throughout the course of HIV infection and during highly active antiretroviral therapy.
The quantification of human immunodeficiency virus type 1 (HIV-1) RNA in plasma has revolutionized the management of infected patients. This parameter is the main prognostic factor for evaluating the progression of the disease and for timing the initiation of treatment and monitoring its efficacy (1, 5, 7, 10, 11, 14, 15, 16). Commercially available assays for the quantification of HIV-1 viral RNA use a variety of techniques, from reverse transcriptase PCR to the branched-DNA assay (2, 12, 20, 21).
The COBAS AMPLICOR HIV-1 MONITOR (CAHIM) test v. 1.5, a quantitative reverse transcriptase PCR assay (3), has been widely used in research as well as in clinical practice. Quantification of HIV-1 RNA with this assay requires two sample preparation procedures: in the first, ultrasensitive method, 500 μl of plasma is centrifuged to increase the input of plasma RNA to achieve a quantitative range of 50 to 2,000 copies/ml of plasma, whereas in the second, standard protocol, with a starting plasma volume of 200 microliters, only an amount of RNA corresponding to 25 microliters of plasma is introduced in the PCR, resulting in a dynamic range of 500 to 500,000 copies/ml. A single-step technique with a wider dynamic range is therefore desirable for routine laboratory applications. The real-time PCR technology already developed for several noncommercial viral assays offers not only broader quantification ranges but also faster turnaround time and more effective prevention of contamination (due to a closed-tube configuration) (4, 13, 23).
In this investigation, we compared the linearity, reproducibility, and sensitivity of the COBAS AMPLICOR HIV-1 MONITOR test v. 1.5 with those of a newly developed real-time PCR assay, the COBAS TaqMan HIV (HPS-CTMHIV) assay, which has a semiautomated format and dedicated hardware and software.
MATERIALS AND METHODS
In this multicenter trial, we evaluated the performance characteristics of the HPS-CTMHIV PCR assay, a semiautomated real-time PCR test based on a dual-labeled hybridization probe targeting the gag region. After manual HIV RNA extraction using the High Pure kit (Roche Diagnostics, Mannheim, Germany), which consists of subsequent steps of lysis and glass particle RNA capture and purification, the TaqMan assay is performed using a dedicated COBAS TaqMan 48 instrument and the results are expressed in numbers of HIV RNA copies/ml. The prevention of carryover contamination and the integrity of the sample are ensured by the use of AmpErase and by an internal control (quantitation standard) (8, 17).
All experiments were performed at sites 1, 2, and 3. The sensitivity, specificity, linearity, and subtype performances were analyzed in comparison with those of the CAHIM, a quantitative PCR assay licensed for diagnostic use (3). The HPS-CTMHIV assay and the CAHIM assay were used in accordance with the manufacturer's instructions included in the test package inserts.
Precision, sensitivity, and specificity.
Intra- and interassay reproducibilities were investigated using an eight-member standard HIV RNA panel with titers ranging from 30 to 5 × 106 copies/ml (AcroMetrix, Benicia, CA). Six replicates of each standard panel member were analyzed on three consecutive days. These experiments were also used to estimate the assay's limit of detection, calculated by probit analysis, while the specificity was assessed by testing 100 samples obtained from healthy donors or patients with HIV-unrelated diseases.
Linearity and dynamic range.
Linearity and dynamic range were investigated using 13 threefold serial dilutions and an undiluted sample of a 5 × 106-copies/ml concentration of the AcroMetrix HIV DNA panel member as well as of a high-titer clinical specimen selected at the trial sites. Each dilution was run in triplicate.
Subtype identification.
The HPS-CTMHIV assay's ability to detect and quantify different subtypes with the same efficiency was analyzed using a standard panel consisting of eight members, representing the group M clade, subtypes A to H, tested in triplicate on three consecutive days (HIV-1 RNA clade performance panel PRD201; Boston Biomedica Inc.).
Comparison of assays.
Parallel experiments were performed at one of the trial sites (site 1) with the CAHIM assay, a quantitative PCR assay with a 4-log dynamic range of concentrations of 50 to 750,000 copies/ml obtained by combining two different sample preparation procedures, the standard and the ultrasensitive (21). In addition, to compare the main performance characteristics of the two systems, we also analyzed their relative performances in quantifying 60 clinical samples taken from patients with HIV infection and different viral loads.
Both the HPS-CTMHIV assay and the CAHIM assay have been approved by the European Union for in vitro diagnostic applications (CE mark) and are being reviewed by the FDA.
RESULTS
In this study, we report the results of a three-site multicenter evaluation of the HPS-CTMHIV assay, a real-time PCR system specifically developed to improve the ease of use, turnaround time, and overall efficacy of current quantitative PCR assays.
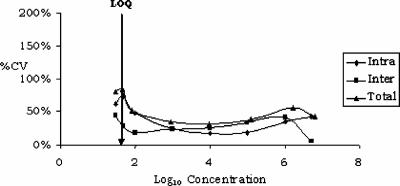
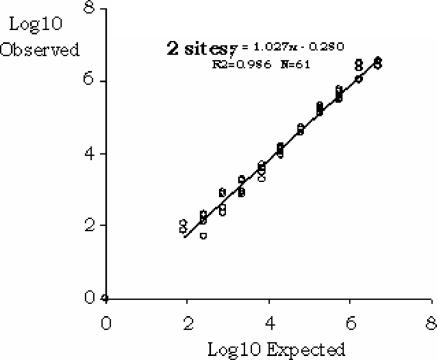
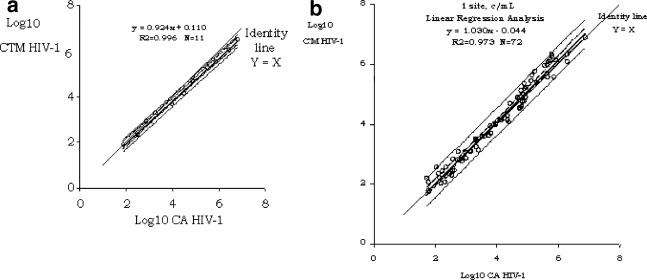
Using an HIV RNA standard panel, we investigated the HPS-CTMHIV assay's precision and detection limit across a range of concentrations from 10 to 107 copies/ml. In several experiments, run on three consecutive days with six replicates of each panel member, we observed a mean total precision (intra- and interassay reproducibility) of 56% (31% to 81% coefficient of variation) and a 95% detection limit of 53 copies/ml as estimated by probit analysis (Fig. 1 and Tables 1 and 2). The assay results were linear over a 5-log range from 10 to 107 copies/ml, and the correlation between the expected and observed results was very high, with an R of 0.98 (Fig. 2). Similar findings were made when data obtained with the HPS-CTMHIV assay were compared to those produced with the CAHIM assay using both the standard and the ultrasensitive procedure. Samples with titers higher than 105 copies/ml had to be further diluted, as they were above the upper limit of the CAHIM assay's reportable range (Fig. 3a). An excellent correlation between the two tests was also noted when we analyzed 72 clinical samples taken from patients with chronic HIV infection and a wide range of viremia levels (Fig. 3b).
FIG. 1.
Intra- and interassay variability of the HPS-CTMHIV assay. Six replicates of an eight-member standard panel were tested by the participating laboratories on three consecutive days. CV, coefficient of variation; LOQ, limit of quantification.
TABLE 1.
Precision of HPS-CTMHIV testa
| Panel member | Concn
|
Mean concn
|
No.b | Precision
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Copies/ml | Log10 | Copies/ml | Log10 | SD (Log10)
|
CV (%)c
|
||||||
| Interassay | Intraassay | Total | Interassay | Intraassay | Total | ||||||
| 1 | 30 | 1.5 | 31 | 1.5 | 10 | 0.18 | 0.25 | 0.31 | 45 | 61 | 81 |
| 2 | 50 | 1.7 | 47 | 1.7 | 34 | 0.12 | 0.25 | 0.28 | 28 | 74 | 84 |
| 3 | 100 | 2.0 | 82 | 1.9 | 47 | 0.08 | 0.20 | 0.21 | 18 | 48 | 52 |
| 4 | 1,000 | 3.0 | 916 | 3.0 | 48 | 0.10 | 0.11 | 0.14 | 23 | 25 | 34 |
| 5 | 10,000 | 4.0 | 9,665 | 4.0 | 47 | 0.11 | 0.07 | 0.13 | 26 | 17 | 31 |
| 6 | 100,000 | 5.0 | 125,050 | 5.1 | 48 | 0.14 | 0.08 | 0.16 | 33 | 17 | 38 |
| 7 | 1,000,000 | 6.0 | 1,621,759 | 6.2 | 48 | 0.16 | 0.14 | 0.22 | 41 | 34 | 56 |
| 8 | 5,000,000 | 6.7 | 6,227,011 | 6.8 | 43 | 0.02 | 0.17 | 0.17 | 5 | 42 | 43 |
Assessed with multiple replicates of an eight-member standard panel of HIV RNA with consecutive runs.
Number of replicates found.
CV, coefficient of variation.
TABLE 2.
Limit of detection as assessed by probit analysisa
| RNA concn (copies/ml) | No. of replicates found | Hit rate (%) |
|---|---|---|
| 0 | 0 | 0 |
| 30 | 36 | 67 |
| 50 | 50 | 93 |
| 100 | 54 | 100 |
| 1,000 | 54 | 100 |
A total of 54 replicates of each member of the standard panel were tested with the HPS-CTMHIV assay. Limit of detection (95% CI), 53.83 (45.13 to 83.62).
FIG. 2.
Linear correlation of observed versus expected results of HPS-CTMHIV test as assessed with 13 threefold serial dilutions and an undiluted sample of a 106-copies/ml concentration of a standard panel member. R2, correlation coefficient.
FIG. 3.
Linear range correlation between the HPS-CTMHIV and the CAHIM tests as assessed with 13 threefold serial dilutions and an undiluted sample of a 106-copies/ml concentration of a standard panel member (a) and with 72 clinical samples (b). R2, correlation coefficient.
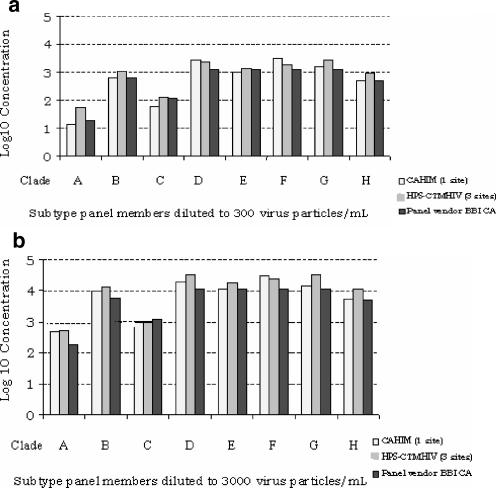
We also used a collection of 100 samples obtained from healthy donors or patients with HIV-unrelated diseases to assess the HPS-CTMHIV assay's specificity. No HIV RNA was detected by either the HPS-CTMHIV or the CAHIM assay, with 100% specificity (data not shown). All major M clade subtypes, including subtypes A to H, were accurately detected and quantified when analyzed at two different concentrations, 300 and 3,000 copies/ml (Fig. 4a and b).
FIG. 4.
Identification of subtypes A to H by the HPS-CTMHIV assay as assessed by analyzing an eight-member standard panel diluted to a nominal concentration of 3 × 102 copies/ml (a) and 3 × 103 copies/ml (b). HIV RNA log concentrations were assessed using the HPS-CTMHIV and the CAHIM tests and compared to the titers calculated by the panel manufacturer (BBI CA, Boston Biomedica Inc., MA).
DISCUSSION
The HPS-CTMHIV test has been developed to standardize the real-time technique based on TaqMan instruments and to improve the performance of the existing PCR assays (13). Based on this evaluation, the HPS-CTMHIV assay appears to have achieved these goals, confirming its expected wide dynamic range and increased sensitivity. Our data indeed show that the assay can quantify concentrations ranging from 10 to 107 copies/ml, thus including and extending the range obtainable by the CAHIM test, the FDA- and CE-approved test we used for system comparison. However, the CAHIM assay makes use of two different sample preparation procedures, the standard and the ultrasensitive. Specimens that are negative by the former, at a detection limit of 400 copies/ml, have to be reevaluated using the latter, which allows for a range of 50 to 5 × 104 copies/ml (21). Although laboratory managers may decide beforehand which preparation to use based on the expected HIV titers of the sample, the possibility of avoiding repeat testing by using the HPS-CTMHIV test instead of the CAHIM test represents a significant advantage in terms of cost effectiveness and workflow.
The HPS-CTMHIV assay's precision, as assessed by intra- and interassay reproducibility, is good and is comparable to that of the CAHIM assay. Further improvement is expected when the RNA extraction procedure, presently labor-intensive and lengthy, is automated (6). Medium-sized to large laboratories, with high sample turnover, may indeed benefit from fully automated sample preparation using dedicated instrumentation (6, 18). In this respect, the COBAS AmpliPrep instrument, a nucleic acid extractor already in use in conjunction with the CAHIM (COBAS AmpliPrep/COBAS AMPLICOR HIV MONITOR) reagents, is being validated as the front-end instrument for the HPS-CTMHIV test (19).
The issue of potential carryover has already been solved for the CAHIM assay through pretreatment with uracyl-N-glycosylase, the enzyme that specifically inactivates dUTP-containing amplicons. This step is also included in the HPS-CTMHIV assay, with additional safety derived from the closed-tube procedure typical of the hydrolysis probe technology used with TaqMan instruments.
This investigation was not designed to evaluate the efficacy of HIV-1 RNA quantification for the various non-B subtypes, since only two dilutions of a single strain corresponding to the most frequent HIV-1 subtypes were tested. However, in this study we show that both tests are able to detect the subtypes tested at low HIV RNA levels (300 copies/ml) and provide similar results at the higher titers tested (3,000 copies/ml). Additional studies of larger numbers of clinical samples are warranted to confirm these data and to assess the ability of the HPS-CTMHIV assay to detect newly emerging strains and recombinants (9, 22).
In conclusion, we have shown that the HPS-CTMHIV test provides performances similar to those of the licensed CAHIM test with regard to specificity, sensitivity, and precision and has additional advantages in terms of a larger dynamic range, high throughput, and absence of carryover during the PCR amplification procedure.
Acknowledgments
COBAS, TaqMan, AmpliPrep, AMPLICOR, High Pure, and AmpErase are trademarks of Roche.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 2.Collins, M. L., B. Irvine, D. Tyner, E. Fine, C. Zayati, C. Chang, T. Horn, D. Ahle, J. Detmer, L. P. Shen, J. Kolberg, S. Bushnell, M. S. Urdea, and D. D. Ho. 1997. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 25:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill, C. E., A. M. Green, J. Ingersoll, K. A. Easley, F. S. Nolte, and A. M. Caliendo. 2004. Assessment of agreement between the AMPLICOR HIV-1 MONITOR test versions 1.0 and 1.5. J. Clin. Microbiol. 42:286-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holland, P. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes, M. D., V. A. Johnson, M. S. Hirsch, J. W. Bremer, T. Elbeik, A. Erice, D. R. Kuritzkes, W. A. Scott, S. A. Spector, N. Basgoz, M. A. Fischl, R. T. D'Aquila, et al. 1997. Monitoring plasma HIV-1 RNA levels in addition to CD4+ lymphocyte count improves assessment of antiretroviral therapeutic response. Ann. Intern. Med. 126:929-938. [DOI] [PubMed] [Google Scholar]
- 6.Jungkind, D. 2001. Automation of laboratory testing for infectious diseases using the polymerase chain reaction—our past, our present, our future. J. Clin. Virol. 20:1-6. [DOI] [PubMed] [Google Scholar]
- 7.Katzenstein, D. A., S. M. Hammer, M. D. Hughes, et al. 1996. The relation of virologic and immunologic markers to clinical outcomes after nucleoside therapy in HIV-infected adults with 200 to 500 CD4 cells per cubic millimeter. N. Engl. J. Med. 335:1091-1098. [DOI] [PubMed] [Google Scholar]
- 8.Longo, M. C., M. S. Berninger, and H. L. Hartley. 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93:125-128. [DOI] [PubMed] [Google Scholar]
- 9.McCutchan, F. E., J. L. Sankale, S. M'Boup, B. Kim, S. Tovanabutra, D. J. Hamel, S. K. Brodine, P. J. Kanki, and D. L. Birx. 2004. HIV type 1 circulating recombinant form CRF09_cpx from west Africa combines subtypes A, F, G, and may share ancestors with CRF02_AG and Z321. AIDS Res. Hum. Retrovir. 20:819-826. [DOI] [PubMed] [Google Scholar]
- 10.Mellors, J. W., A. Munoz, J. V. Giorgi, J. B. Margolick, C. J. Tassoni, P. Gupta, L. A. Kingsley, J. A. Todd, A. J. Saah, R. Detels, J. P. Phair, and C. R. Rinaldo, Jr. 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 126:946-954. [DOI] [PubMed] [Google Scholar]
- 11.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 12.Mulder, J., N. McKinney, C. Christopherson, J. Sninsky, L. Greenfield, and S. Kwok. 1994. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J. Clin. Microbiol. 32:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niesters, H. B. 2004. Molecular and diagnostic clinical virology in real time. Clin. Microbiol. Infect. 10:5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Brien, W. A., P. M. Hartigan, E. S. Daar, M. S. Simberkoff, J. D. Hamilton, et al. 1997. Changes in plasma HIV RNA levels and CD4+ lymphocyte counts predict both response to antiretroviral therapy and therapeutic failure. Ann. Intern. Med. 126:939-945. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien, W. A., P. M. Hartigan, D. Martin, et al. 1996. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N. Engl. J. Med. 334:426-431. [DOI] [PubMed] [Google Scholar]
- 16.Riddler, S. A., and J. W. Mellors. 1997. HIV-1 viral dynamics and viral load measurement: implications for therapy. AIDS Clin. Rev. 1997-1998:47-65. [PubMed] [Google Scholar]
- 17.Rosenstraus, M., Z. Wang, S.-Y. Chang, D. DeBonville, and J. P. Spadoro. 1998. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J. Clin. Microbiol. 36:191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schorling, S., G. Schalasta, G. Enders, and M. Zauke. 2004. Quantification of parvovirus B19 DNA using COBAS AmpliPrep automated sample preparation and LightCycler real-time PCR. J. Mol. Diagn. 6:37-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stelzl, E., A. Kormann-Klement, J. Haas, E. Daghofer, B. I. Santner, E. Marth, and H. H. Kessler. 2002. Evaluation of an automated sample preparation protocol for quantitative detection of hepatitis C virus RNA. J. Clin. Microbiol. 40:1447-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevens, W., T. Wiggill, P. Horsfield, L. Coetzee, and L. E. Scott. 2005. Evaluation of the NucliSens EasyQ assay in HIV-1-infected individuals in South Africa. J. Virol. Methods 124:105-110. [DOI] [PubMed] [Google Scholar]
- 21.Sun, R., J. Ku, H. Jayakar, J.-C. Kuo, D. Brambilla, S. Herman, M. Rosenstraus, and J. Spadoro. 1998. Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 36:2964-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong, C. Y. W., J. Mullen, R. Kulasegaram, A. De Ruiter, S. O'Shea, and I. L. Chrystie. 2005. Genotyping of B and non-B subtypes of human immunodeficiency virus type 1. J. Clin. Microbiol. 43:4623-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss, J., H. Wu, B. Farrenkopf, T. Schultz, G. Song, S. Shah, and J. Siegel. 2004. Real time TaqMan PCR detection and quantitation of HBV genotypes A-G with the use of an internal quantitation standard. J. Clin. Virol. 30:86-93. [DOI] [PubMed] [Google Scholar]