Figure 1.
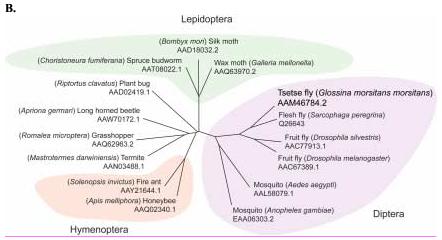
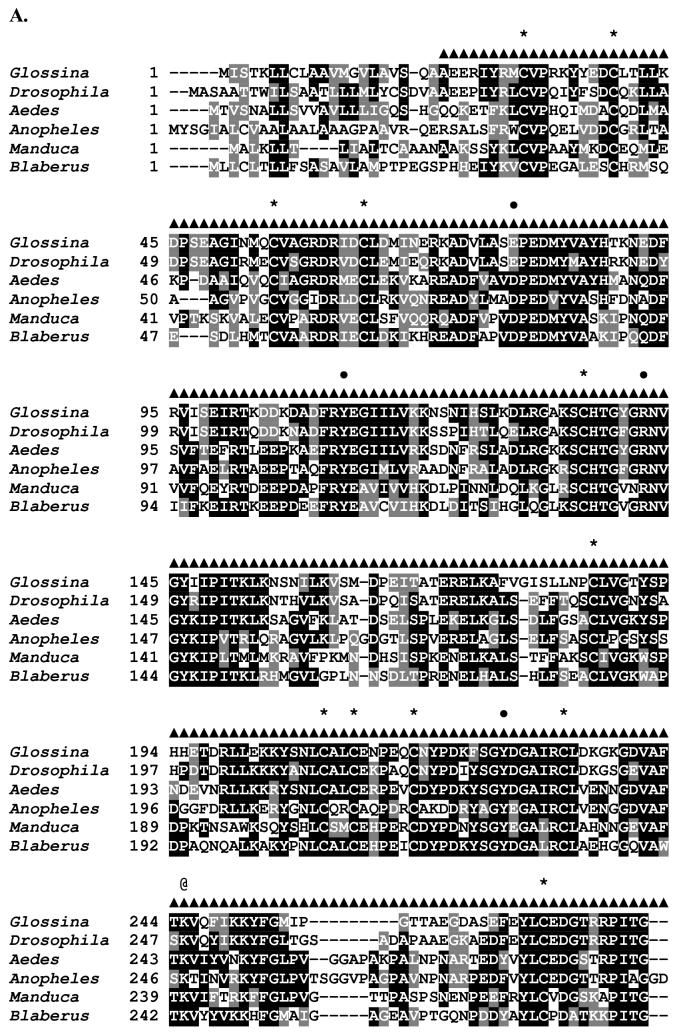
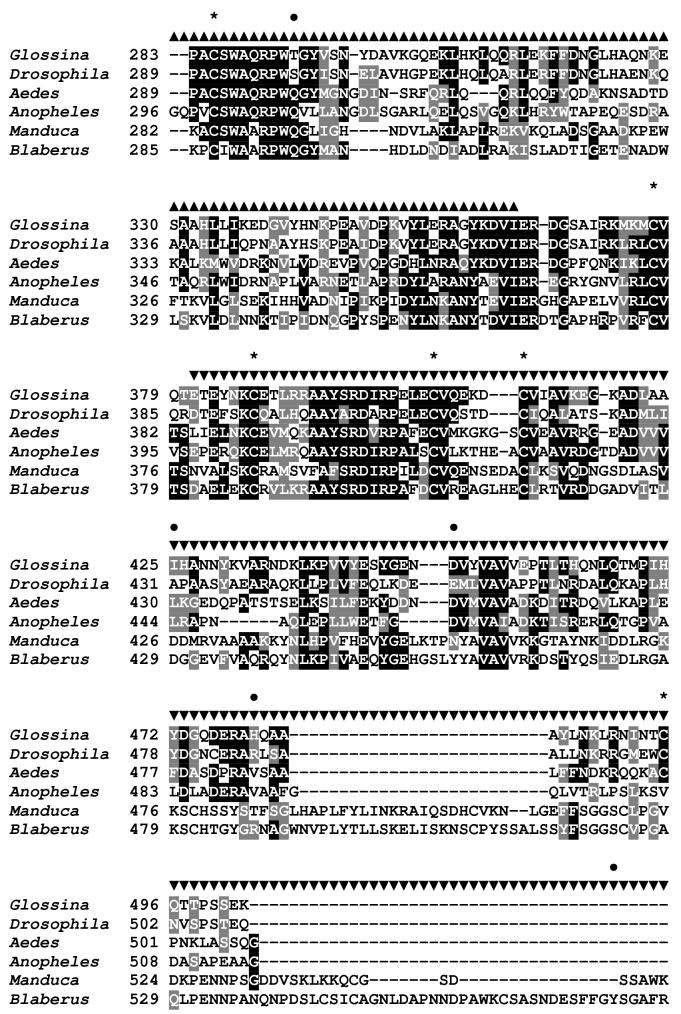
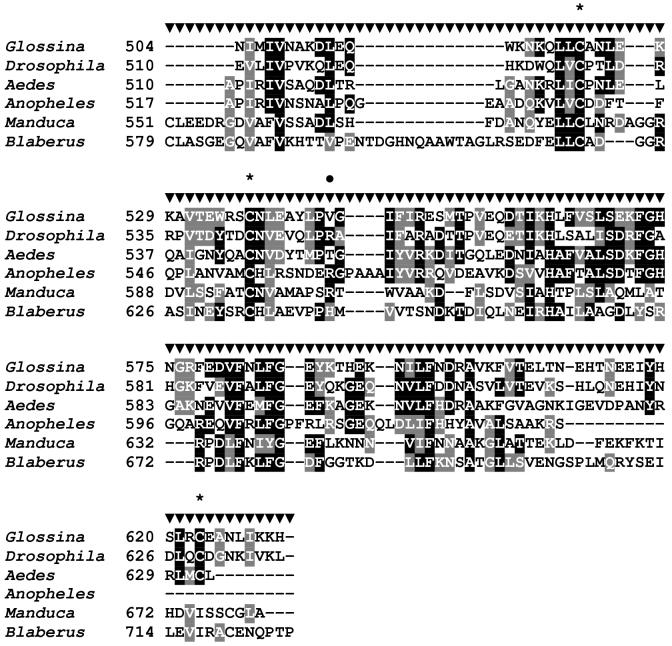
Sequence analysis of the G. m. morsitans transferrin homologue (GmmTsf1).
(A) Multiple alignment of transferrin N-terminal lobe amino acid sequences from the following: G. m. morsitans; D. melanogaster (Genbank Accession: CG6186); Ae. aegypti (AAB87414), An. gambiae (EAA06303), M. sexta (AAA29338), and B. discoidalis (AAA27820). The output for multiple sequence alignments is based on a ClustalW consensus sequence and indicates ≥50% identity (black shading) or ≥50% similarity (gray shading). Amino acid residues necessary for iron binding in vertebrate transferrins are indicated by a ‘•’, a conserved lysine with@, and conserved cysteine residues by ‘*”. (B) Molecular phylogeny of dipteran transferrins. Pairwise alignment and tree generation for GmmTsf1 was performed using the CLUSTALW program (http://clustalw.genome.jp/). Accession numbers for sequences used in the analysis have been included.