FIG. 5.
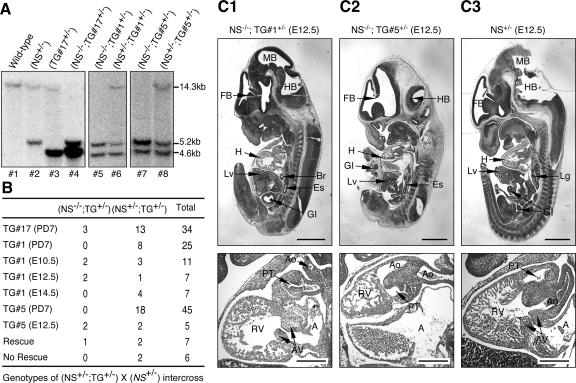
NS transgene rescued homozygous NS-null (NS−/−) embryos in a dose-dependent manner. (A) To rescue NS−/− mice using the transgenic alleles, compound heterozygous mice (NS+/− TG+/−) were crossed with NS+/− mice. The genotypes of their bigenic offspring were determined by Southern blots in which the NS transgene, NS-null, and endogenous NS alleles would yield 4.6-kb, 5.2-kb, and 14.3-kb fragments, respectively. Pups 4, 5, and 7 represented NS−/− TG+/− mice or embryos. The transgene copy numbers, determined by the intensity ratios between the transgene and the NS-null allele, were 2, 1, and 4 for the TG#1, TG#5, and TG#17 lines, respectively. (B) The high-expression TG#17 line could functionally rescue NS−/− embryos into adulthood. By contrast, the lower-copy-number TG#1 could rescue the NS-null embryos only up to E12.5. The rescue probability of the TG#1 allele dropped significantly between E12.5 and E14.5. Although identifiable at E12.5, NS−/− TG#5+/− embryos appeared developmentally delayed. The predicted ratios for rescue and no rescue are shown. (C) Hematoxylin-eosin immunohistochemistry of the sagittal sections of the whole NS−/− TG#1+/−, NS−/− TG#5+/−, and NS+/− embryos at E12.5 are shown in the top panels of C1, C2, and C3, respectively. Higher magnifications of the heart are shown in the bottom panels. The outflow track (pulmonary trunk and aorta) and the right ventricle of the NS−/− TG#1+/− embryo were underdeveloped. In addition to dilated atria and small ventricles, the pulmonary trunk and aorta of the NS−/− TG#5+/− embryo were in the wrong position, indicating an outflow track-remodeling defect. Abbreviations: FB, forebrain; MB, midbrain; HB, hindbrain; H, heart; Lv, liver; Lg, lung; Br, bronchus; Es, esophagus; GI, gastrointestinal tract; RV, right ventricle; A, atrium; PT, pulmonary trunk; Ao, aorta; AV, atrioventricular cushion. Bars: 1 mm (top panels), 300 μm (bottom panels).