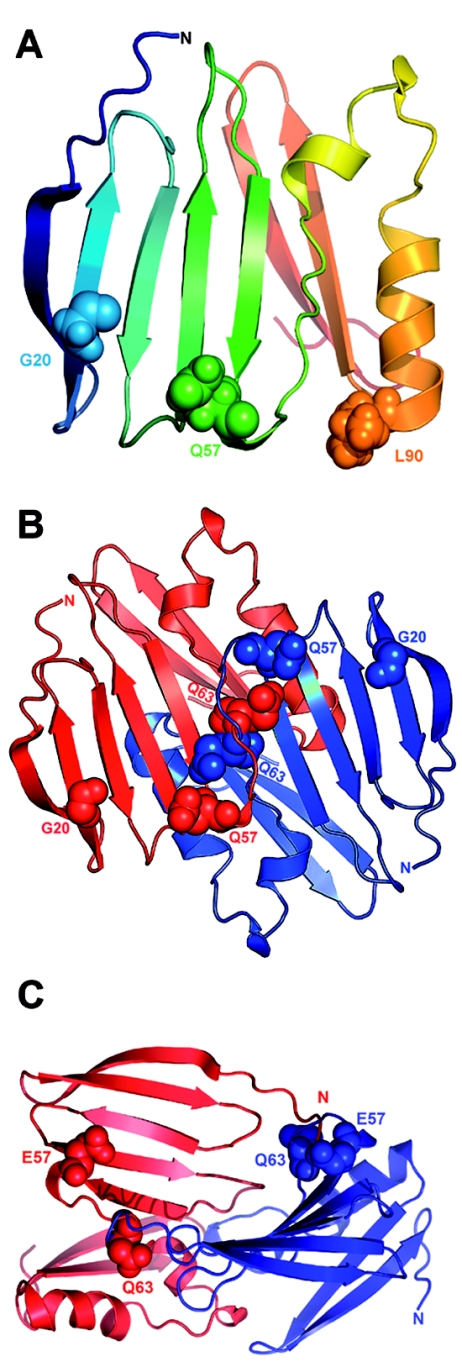
Figure 5. .
Three-dimensional structural models of claudin-19. A, The first 115 aa of claudin-19 are shown as a ribbon diagram in a gradient of rainbow colors, with the N terminus depicted in blue. The G20D mutation is located in the first transmembrane region that is buried in the membrane. The strong change in the charge pattern by the replacement of glycine with aspartic acid presumably influences intramolecular interactions that will alter the signal sequence. The Q57E mutation is located in the first extracellular loop and is likely to affect electrostatic interactions that result in altered protein-protein interactions. The L90P mutation is located in the second transmembrane domain. B, Potential model of claudin-19 homodimer formation. A representative docking solution—which shows the involvement of the first extracellular loop in the dimer formation with Q57 of the first molecule interacting with Q61 and Q63 of the second molecule—is depicted. The two claudin-19 molecules are colored in red and blue. The claudin-19 homodimer seems to be stabilized by interaction of Q57 of one molecule with Q63 of the partner molecule. C, Effect of the Q57E mutation on claudin-19 homodimer formation. Docking calculations were repeated after performing in silico mutation of Q57E in the monomeric model. The dimer formation was disrupted with the mutant protein, possibly because of strong electrostatic repulsion from opposing E residues.