Figure 1. .
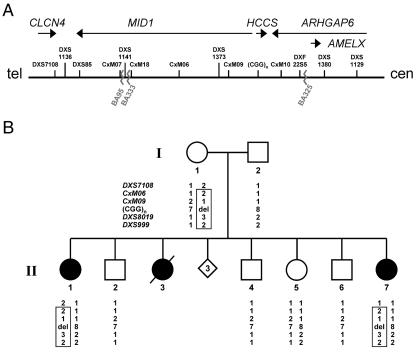
Minimal MLS critical region and haplotype analysis of the family of patient II.7. A, Schematic representation of the minimal MLS critical region (610 kb) in Xp22.2, defined by the breakpoints of patients BA333 and BA325 (jagged lines). The figure is not drawn to scale. The uppermost arrows show the orientation of genes within the critical region. The position of DNA marker loci previously mapped in this region is shown. B, Haplotype analysis of six representative polymorphic markers in Xp22.2-p22.13 in the family of patient II.7. Whereas markers DXS7108, CxM06, CxM09, and SNP rs5901444—(CGG)n—are positioned in the MLS critical region and indicated in panel A, loci DXS8019 and DXS999 are located centromeric to the critical region. The polymorphic (CGG)n repeat is located in the HCCS 5′ UTR, and the respective repeat length is given. Alleles are shown below the pedigree symbols. del = Deletion of one allele. The haplotype shared by the affected sisters (II.1 and II.7) and their mother (I.1) is boxed. Both patients II.7 and II.1 carry only the paternal allele, with eight CGG repeats, whereas the unaffected sister (II.5) is heterozygous for the trinucleotide repeat. Segregation analysis showed that the three brothers (II.2, II.4, and II.6) and the unaffected sister (II.5) carry a different maternal haplotype than do II.1 and II.7.