Figure 3. .
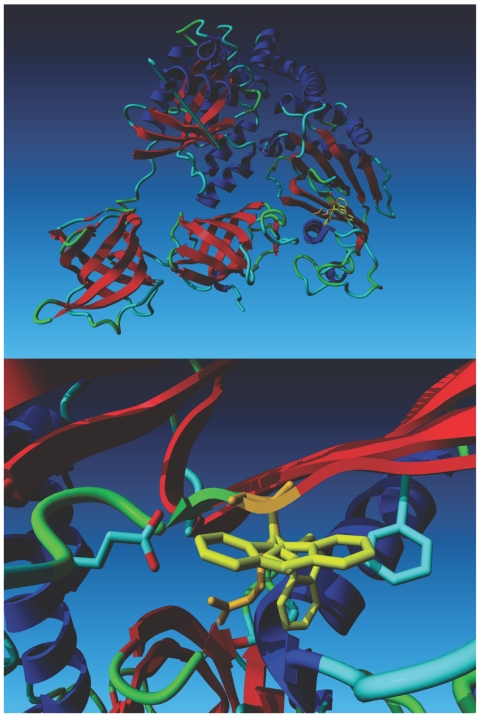
Computational modeling based on the crystal structure of the bovine EFTu•EFTs complex. The top panel shows the structure of the Tu•Ts dimer. Tu is on the left side of the model, and Ts is on the right. The wild-type Arg 333 residue (depicted in orange) is superimposed on three Trp rotamers (depicted in yellow). The lower panel is a close-up of the region containing the mutation, which shows that the Arg residue fits well in front of the C-terminal side of the helix beneath it. The most favorable Trp rotamer bumps into this same helix. The second- and third-best local minima for the Trp rotamer bump into Glu 644 and Phe 637, respectively. The second rotamer also bumps into its own backbone.