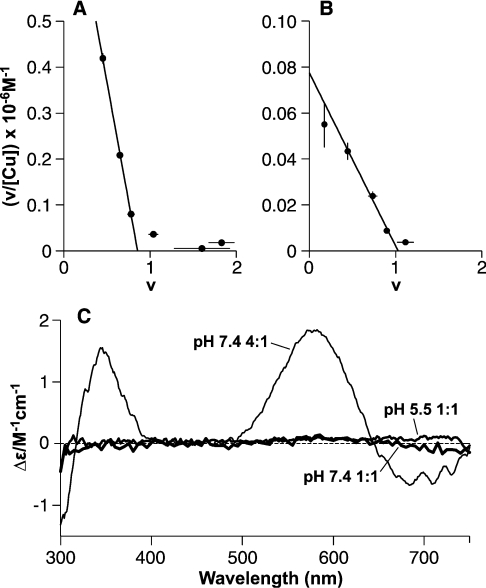
Figure 2. A 1:1 Cu(PrP57–91) complex is revealed by equilibrium dialysis with a mode of binding distinct from that in the Cu4(PrP57–91) complex.
Equilibrium dialysis of PrP57–91 shows that the peptide binds a single copper (II) ion with greater affinity than subsequent ones. Scatchard plots show the fractional occupancy (v) against v/[Cufree], with the slope of the fitted lines giving the apparent association constant and the intercept on the x-axis giving the number of sites of this affinity. (A) Dialysis against CuSO4 in 5 mM Tris buffer, pH 7.4, reveals a single high-affinity site (intercept=0.86) with an apparent dissociation constant of 1.0±0.2 μM. Binding at a second site was observed under these conditions, but is substantially weaker. (B) A single-high affinity site (intercept=1.03) is also present at pH 5.5 in 10 mM acetate buffer, with an apparent dissociation constant of 13±2 μM. (C) CD spectra show that these 1:1 complexes involve a different form of co-ordination to that seen in the 4:1 complex. The spectra of the 1:1 complexes formed in 5 mM Tris buffer at pH 7.4 (15 μM PrP57–91 and 25 μM CuSO4, i.e. 1:1 CuSO4 and 10 μM free CuSO4) and in 10 mM acetate of PrP57–91 pH 5.5 (42 μM PrP57–91 and 300 μM CuSO4, i.e. 1:1 CuSO4 and 240 μM free CuSO4) do not give any measurable CD bands, in contrast with the 4:1 complex formed in NEM buffer at pH 7.4.