Abstract
The sel-6 gene was previously identified in a screen for suppressors of the egg-laying defect associated with hypermorphic alleles of lin-12 (Tax et al. 1997). Here we show that sel-6 and two other previously defined genes, mal-2 and emb-4, are the same gene, now called “emb-4.” We perform a genetic and molecular characterization of emb-4 and show that it functions cell autonomously as a positive regulator of lin-12 activity. Viable alleles identified as suppressors of lin-12 are partial loss-of-function mutations, whereas the null phenotype encompasses a range of lethal terminal phenotypes that apparently are not related to loss of lin-12/Notch signaling. emb-4 encodes a large nuclearly localized protein containing a predicted ATPase domain and has apparent orthologs in fission yeast, plants, and animals.
RECEPTORS of the LIN-12/Notch family mediate cell–cell interactions that specify cell fate during animal development and have an unusual mechanism of signal transduction (Greenwald 2005). LIN-12/Notch proteins are type I transmembrane proteins that undergo modifications upon their transit to the cell surface. Binding of a protein of the Delta-Serrate-LAG-2 family triggers proteolytic cleavage in the extracellular domain, resulting in ectodomain shedding. This extracellular cleavage is mediated by a protease of the ADAM family and creates a truncated form with a short extracellular domain. This truncated form serves as the substrate for a protease complex that includes presenilin, which cleaves within the transmembrane domain. Transmembrane cleavage releases the intracellular domain, which translocates to the nucleus and forms a transcriptional activation complex with the sequence-specific DNA-binding protein LAG-1 (Suppressor of Hairless in Drosophila and CBF1/RBP-J in mammals) and other cofactors. Thus, LIN-12/Notch proteins are essentially membrane-tethered transcription factors that are released upon ligand binding.
The identification of trans-acting factors that constitute the core machinery of signal transduction or modulate the activity of the signaling pathway has been fruitfully approached using genetics in Caenorhabditis elegans. The main approach has been to isolate extragenic suppressors of phenotypes caused by elevated or reduced lin-12 activity. Genes thereby defined—many named sel genes, for suppressor/enhancer of lin-12—include those that encode proteases involved in cleaving LIN-12/Notch, components of the transcription activation complex, and factors that affect LIN-12 stability or trafficking. Many of these proteins are conserved and play similar roles in other animals (reviewed in Greenwald 2005).
The sel-6 gene was first identified in a screen for suppressors of a constitutively active allele of lin-12 (Tax et al. 1997). Our additional genetic analysis indicates that sel-6 functions as a positive regulator of lin-12 activity. The alleles of sel-6 that were recovered in screens for sel genes appear to be hypomorphic. During the course of investigating sel-6, we found that alleles had been recovered in two independent screens for maternal-effect essential genes and had been assigned to two genes, emb-4 (Miwa et al. 1980; Schierenberg et al. 1980) and mal-2 (Hekimi et al. 1995). As emb-4 was the first published name, it has become the official name of this gene. We show that emb-4 encodes a protein with a putative ATPase domain with apparent orthologs in higher eukaryotes and that it affects lin-12 activity in a cell-autonomous manner. We discuss possible mechanisms by which emb-4 may influence lin-12 activity. In an accompanying article in this issue, Checchi and Kelly (2006) discuss the role of emb-4 in a mechanism that keeps germ cell precursors transcriptionally quiescent.
MATERIALS AND METHODS
Strains:
Experiments were performed at 20° unless indicated otherwise. The wild-type parent for all strains was C. elegans var. Bristol N2 (Brenner 1974), except for mapping experiments using GS3063, a hybrid strain in which lin-12(n302) was placed into the Hawaiian strain CB4856 background by six backcrosses (data not shown). GS3063 [lin-12(n302); him-5(e1467)] has all the single-nucleotide polymorphisms (SNPs) that we tested on LG V.
The main alleles used in this study are the following: LG III, lin-12(n302), lin-12(n950), lin-12(n379), lin-12(n941) (Greenwald et al. 1983; Greenwald and Seydoux 1990), lin-12(ar170) (Hubbard et al. 1996), and glp-1(ar202) (Pepper et al. 2003); and LG V, sel-6(sa44, n1256) (Tax et al. 1997), mal-2(qm31) (Hekimi et al. 1995), and emb-4(hc60) (Miwa et al. 1980). Additional information about these alleles and markers used for mapping and for facilitating genetic analysis can be found via Wormbase at http://www.wormbase.org.
arIs53[lin-12(ΔE)∷gfp] expresses a GFP-tagged version of LIN-12 lacking most of its extracellular domain, a constitutively active form of the receptor (Shaye and Greenwald 2005).
We used transgene arIs51[cdh-3∷gfp] to facilitate visualization of anchor cells (Karp and Greenwald 2003).
Mutant analysis and scoring:
For scoring the egg-laying defective (Egl) phenotype, L4 larvae were picked onto separate plates and scored 2 days later. An animal was scored as non-Egl if it laid eggs and as Egl if it did not lay eggs or if it formed a bag of worms. The egg-laying potential of sterile animals could not be assessed.
To assess the number of anchor cells (ACs), L3 larvae were scored by Nomarski optics and ACs were identified by morphology and GFP expression using the marker arIs51[cdh-3∷gfp]. For the Multivulva (Muv) phenotype, L4 larvae were generally picked onto separate plates and checked for the number of pseudovulvae the next day.
For germline proliferation, L1 larvae of hermaphrodites carrying glp-1(ar202) were transferred to individual plates at 25° and scored for progeny production.
Fine-scale genetic mapping of sel-6(sa44):
We used GS3063, a lin-12(n302)-containing derivative of the Hawaiian strain CB4856, for further mapping to facilitate recombinant analysis.
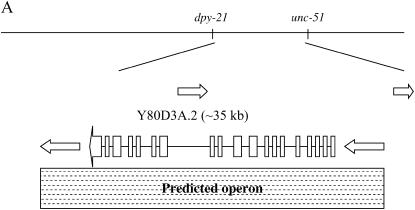
lin-12(n302); sel-6(sa44) unc-51(e369) rol-9(sc148) hermaphrodites were mated with GS3063 males and non-Unc, non-Rol F2's were recovered. Homozygosity for sel-6 was determined by cloning 10 F3 progeny from each recombinant candidate and checking that all segregated egg-laying-competent animals. A total of 78 Sel non-Unc non-Rol recombinants were obtained. Of the 78 recombinants, 4 placed sel-6 to the left of a newly identified C-to-T SNP, ik1, between genes Y80D3A.10 and Y80D3A.5 (using the primer pair ik1F: 5′-CACAGGTGCTCAAAATCACAA-3′ and ik1R:5′-TGGCGCAATTCTCGATGTTGC-3′). This SNP introduces an XbaI restriction site.
GS3063 males were mated with lin-12(n302); dpy-21(e428) sel-6(sa44) hermaphrodites. Non-Dpy F2's were picked from F1 cross-progeny. We assessed the presence and homozygosity of sel-6(sa44) as above. A total of 79 Sel non-Dpy recombinants were obtained. Of the 79 recombinants, 1 placed sel-6 to the right of a newly identified C-to-A SNP, ik2, between genes Y17D7A.2 and Y80D3A.1 (using the primer pair ik2F: 5′-AAACCAGGAAAGGGGCGGAGC-3′ and ik2R: 5′-AGAGCATCGCGAAAAAGGTGG-3′). This SNP introduces a DraI restriction site.
Combined SNP mapping data delineate the region of sel-6 between the predicted genes Y17D7A.2 and Y80D3A.5, containing five predicted genes.
Phenocopy of lin-12(n302) suppression by RNA-mediated interference against predicted genes in the sel-6 interval and sequencing:
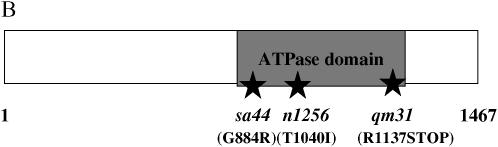
We constructed plasmids expressing double-stranded RNA (dsRNA) in the vector pPD129.36 (Timmons et al. 2001), corresponding to four of the five genes in the sel-6 mapping interval (for construct details, see below). These constructs were transformed into bacterial strain HT115 (Timmons and Fire 1998; Timmons et al. 2001). lin-12(n302) adults were placed on lawns of such bacteria and their progeny were scored for egg-laying ability. Some eggs were observed upon feeding with pIK21 (corresponding to Y80D3A.2) and pIK24 (corresponding to Y80D3A.3). Y80D3A.3 is contained within intron 12 of Y80D3A.2. These two predicted genes were sequenced using DNA from sel-6(sa44) and sel-6(n1256) backgrounds. Single-base-pair changes were found within the coding region of Y80D3A.2 in both cases. The PCR product obtained using one pair of primers, Y80D3A.2(7F) (5′-TAGTCCTAAGCTCTCCAGGTG-3′) and Y80D3A.2(7R) (5′-AAAATCCTCAATTTTCCGGGG-3′), contains a G-to-A mutation in sel-6(sa44) (G884R). The PCR product obtained using a different pair of primers, Y80D3A.2(10F) (5′-AAATTTTAGAGTTTTTCGACG-3′) and Y80D3A.2(10R) (5′-CCAAATTTCAGCAAATTCAGC-3′), contains a C-to-T mutation in sel-6(n1256) (T1040I). sa44 is in exon 10, and n1256 is in exon 13 of Y80D3A.2. Y80D3A.4 and Y80D3A.10 were eventually also sequenced to confirm that they did not contain mutations in sel-6 mutant strains. Two additional noncomplementing mutations have been sequenced and found to be in exons of Y80D3A.2 (P. Checchi, personal communication).
cDNA analysis:
yk1132d03 was the only cDNA available that appeared to be full-length. It meets the following criteria for a full-length cDNA: (1) it starts from the consensus 3′ splice site for cis- and trans-splicing; (2) a partial spliced-leader sequence is present, but it is not possible to determine whether it is SL1, SL2, or another variant; (3) the ATG translation start site is 8 bp downstream from the 3′ splice site and is preceded by two A residues; and (4) the cDNA ends in a poly(A) sequence. All exon junctions were examined in terms of their genomic context and none appear to result from amplification mistakes. Our analysis does not exclude the possibility that there are alternative splice isoforms, but we found no evidence of it in the available cDNA clones. The full-length SEL-6 protein has 1467 amino acids encoded by 19 exons.
RNA interference assays:
We assessed whether the null phenotype of sel-6 was indeed embryonic and larval lethality as reported by Kamath et al. (2003). We used the two protocols below; only protocol II yielded a high percentage of lethality.
Protocol I:
Either three to five young N2 adults or ∼100 bleached eggs were placed on lawns of bacteria expressing the desired RNA interference (RNAi) constructs against sel-6 and controls at 20°, room temperature, or 25°. The progeny of these worms were observed for occurrence of any embryonic or larval lethality.
Protocol II:
Between 10 and 15 L3–L4 N2 worms were placed on lawns of bacteria expressing the desired RNAi constructs against sel-6 and controls and left at 15° for 72 hr. Three animals, now adults, were removed and placed onto individual plates and allowed to lay eggs for 24 hr at 20°, room temperature, or 25°. The three worms were then removed. The phenotypes of worms that hatched from these eggs (F1) as well as their progeny (F2) were then examined for embryonic and larval lethality (Kamath et al. 2003).
Complementation tests with emb-4(hc60) and mal-2(qm31) and sequencing the mal-2(qm31) allele:
The emb-4 or mal-2 mutant phenotypes include lethality and ectopic growths reminiscent of those observed upon sel-6 RNAi (Cassada et al. 1981; Hekimi et al. 1995). We performed complementation tests as follows: emb-4(hc60) progeny of emb-4(hc60) segregate 100% dead eggs at 25°. emb-4(hc60)/sel-6(sa44) unc-51(e369) were allowed to lay eggs at 15°. A total of 60 of their non-Unc progeny were picked to individual plates as L4 larvae, switched to 25°, and scored for segregation of live progeny. About one-third of these worms segregated only dead eggs; the rest segregated both Unc and non-Unc live worms and occasional dead eggs. emb-4(hc60)/unc-51(e369) were scored in parallel as a negative control and showed the same segregation patterns. sel-6(sa44) thus complements the maternal-effect lethality of emb-4(hc60).
mal-2(qm31) self-progeny of mal-2(qm31) mothers segregate a high percentage of embryonic and larval lethals at 20°. However, we observed no difference in lethality between progeny of mal-2(qm31)/sel-6(sa44) and mal-2(qm31)/+ animals (4/231 eggs laid failed to hatch vs. 2/420). This indicates complementation between sel-6(sa44) and mal-2(qm31) for maternal-effect embryonic lethality.
We sequenced the predicted coding region of Y80D3A.2 in the mal-2(qm31) background. PCR product using the primer pair Y80D3A.2(10F) (5′-AAATTTTAGAGTTTTTCGACG-3′) and Y80D3A.2(10R) (5′-CCAAATTTCAGCAAATTCAGC-3′) contains a C-to-T mutation in mal-2(qm31) (R1137STOP).
Plasmid construction:
For RNAi constructs corresponding to four genes in the sel-6 interval, a PCR product was amplified from N2 genomic DNA within each gene, TOPO-cloned, and then subcloned between XhoI and SacI sites (for pIK21–pIK23) or EcoRV sites (for pIK24) of pPD129.36. The following PCR primers were used: for Y80D3A.2, pIK21F (5′-AGTACTTGGAGGCCTATTTATGG-3′) and pIK21R (5′-GCTCCAAACTGTTCAAAACTCG-3′); for Y80D3A.4, pIK22F (5′-GATCACCCCATTCTCTTCTTC-3′) and pIK22R (5′-TTATAGCACCGTTCCAGACG-3′); for Y80D3A.10, pIK23F (5′-GAGAGTGCAAGTAGTCACAC-3′) and pIK23R (5′-CAATTTTGGGTCTCACCACG-3′); and for Y80D3A3.3, pIK24F (5′-GCAATTAACAGCTCCAAGAAG-3′) and pIK24R (5′-AACATGCTTCTGGTCCTCCTC-3′). The RNAi construct containing the full-length cDNA of Y80D3A.2 (pIK25) was obtained by digesting the yk1132d03 cDNA plasmid with Eco0109I and XbaI and ligating the insert into the corresponding sites in the vector pPD129.36 (Timmons et al. 2001).
The vector PIN2 (D. Levitan and I. Greenwald, unpublished observations) drives inserted sequences under the control of sel-12 regulatory sequences. It contains a unique NotI site inserted at the second amino acid of a sel-12-rescuing genomic fragment. To make pIK33, PIN2∷emb-4cDNA, we first inserted a NotI site in front of the ATG of the cDNA by amplifying a PCR product from the cDNA using the following pair of primers: NruF (5′-CAACAAGCGGCCGCTATGGTGACTAAACGTCATCAAG-3′, where the NotI site is underlined and the ATG of emb-4 is in boldface type) and NruR (5′-CAATCTAGACGCTCTTCCTGCAACGAAAGCTGC-3′, with the added XbaI site underlined). This piece was cloned into the NotI and XbaI sites of pPD129.36. The resulting plasmid was then cut with NruI and a NruI fragment from pIK25 was subcloned into it, yielding pIK30. pIK32 was constructed by ligating the RsrII/XbaI fragment of pIK25 into pIK30. This XbaI site is 3′ to the second NotI site in IK31 that follows the emb-4 cDNA poly(A) sequence. pIK33 was finally obtained by cloning the NotI insert containing the entire emb-4 cDNA into the NotI site of PIN2. All joints and correct orientation of the cDNA were confirmed by sequencing.
To construct pIK56, lin-31p∷gfp∷emb-4cDNA, we first used the PCR fusion approach to generate the N-terminally tagged EMB-4 (Hobert 2002) using a KSGFPS65T vector and a vector containing the emb-4 cDNA subcloned into NotI sites. The relevant primers were A*big (5′-CAACAAGCGGCCGCAAAAATGAGTAAAGGAGAAGAAC-3′), Bbig (5′-CTTGATGACGTTTAGTCACCATTTTGTATAGTTCATCCATGC-3′), and D*big (5′-TACTTCATTGGGCTCACAAGCGGC-3′), where the NotI site is underlined. The PCR product was TOPO cloned and subsequently subcloned into the NotI sites of the plasmid pB253 (Tan et al. 1998).
Worm transformation and rescue experiments:
DNA was micro-injected into the germline of C. elegans. pIK33, PIN2∷emb-4cDNA, was injected at different concentrations together with the co-injection marker ceh-22∷gfp. Many different combinations of micro-injection mixes and recipient strains yielded only eight lines. Lines were obtained only when the concentration of pIK33 was no >2.5 ng/μl. For rescue experiments, the transgenes were placed into appropriate genetic backgrounds. Egg laying was scored in the lin-12(n302); emb-4(sa44) background and the number of pseudovulvae was scored in the arIs53[lin-12(ΔE)∷gfp]; emb-4(sa44) background. No rescue was observed in either background with any of the transgenic arrays. All of the arrays were transmitted at a very low frequency, suggesting that overexpression of emb-4 may be toxic to animals.
Subcellular localization:
pIK56, lin-31p∷gfp∷emb-4cDNA, was micro-injected into the germline of C. elegans at 20 ng/μl together with the co-injection marker ceh-22∷gfp. Three lines were obtained, but no GFP expression was observed. Two lines were then analyzed by immunofluorescence. For immunofluorescence, transgenic animals were fixed according to the modified Finney–Ruvkun procedure and stained with a polyclonal rabbit anti-GFP antibody (Molecular Probes, Eugene, OR) at a 1:300 dilution and a mouse anti-MH27 (Priess and Hirsh 1986) at a 1:10 dilution in antibody buffer A. Secondary antibodies used were goat-anti-rabbit conjugated to Cy2, and goat-anti-mouse conjugated to Cy3, both at 1:300 (Jackson ImmunoResearch, West Grove, PA, nos. 111225144 and 115165146, respectively). The stained animals were mounted onto an agarose pad with a drop of SlowFade (Molecular Probes, no. S-7461) and visualized with a Zeiss Axio Imager Z1 microscope.
Laser microsurgery:
The nucleus of Z4 was ablated with a laser microbeam in newly hatched L1 larvae. Success of ablations was confirmed and the presence of an AC was scored during the late L3 stage.
RESULTS
Molecular identification of sel-6:
sel-6 had been mapped previously to the right arm of LG V (Tax et al. 1997). We mapped the reference allele sel-6(sa44) to a region containing five predicted genes (Figure 1; see materials and methods). We provisionally identified Y80D3A.2 as sel-6 by feeding RNAi (Timmons et al. 2001) using protocol I (see materials and methods) on the basis of the presumption that loss of gene activity would confer a Sel phenotype, i.e., suppression of the 0 AC–Egl defect of lin-12(n302).
Figure 1.—
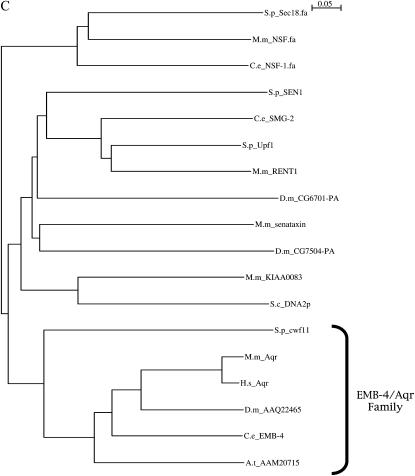
Molecular cloning and DNA sequence analysis of emb-4. (A) Mapping and cloning of emb-4. emb-4 was genetically mapped to the region including the five genes depicted schematically. Y80D3A.2, the second gene in the predicted operon, is emb-4. (B) Schematic of the EMB-4 protein. The ATPase domain has similarity to DEAD-box helicases and AAA ATPases. (C) Phylogenetic analysis. The predicted C. elegans EMB-4 sequence was compared to predicted proteins in Saccharomyces cerevisiae, S. pombe, A. thaliana, C. elegans, Mus musculus and Homo sapiens using NCBI BLAST (http://www.ncbi.nih.gov/BLAST/) (Altschul et al. 1997; Schaeffer et al. 2001). Sequences showing significant alignment were downloaded in FastA format, and the labels for each protein were modified to include a code for species and sequence identifier. Many duplicate entries were eliminated by identifying identical sequences. ClustalX was then used to align the sequences. The orthologs of EMB-4 shown are S. pombe cwf11p (SPBC646), A. thaliana AAM20715, Drosophila melanogaster AAQ22465, M. musculus Aquarius (BAC 65592), and H. sapiens Aquarius (BAA25486). N-ethylmaleimide-sensitive factor family proteins were included as outliers because of their distant similarity to the apparent EMB-4 orthologs.
We did not perform a conventional rescue experiment, as the genomic region is incompletely covered by cosmids and Y80D3A.2 is the second gene in the predicted three-gene operon that spans 45 kb. However, when we sequenced the exons of Y80D3A.2 from sel-6(sa44) and sel-6(n1256) mutants, we observed single-base-pair changes within the coding region of this gene, corroborating the RNAi evidence that Y80D3A.2 is indeed sel-6 (Figure 1). The sequence of putative null, noncomplementing mutations provides further corroboration. We will describe features of the predicted protein product below.
sel-6, mal-2, and emb-4 are a single gene:
When N2 hermaphrodites were fed using protocol II with bacteria expressing double-stranded RNA corresponding to the full-length Y80D3A.2 cDNA, only 8% (8/96) hatching was observed, as compared to 98% (138/141) observed with an empty vector construct. Embryonic lethality was also observed upon dsRNA injection (data not shown). Y80D3A.2(RNAi) viable escapers often displayed unusual outgrowths on the head or the tail of the larvae. The lethality and the presence of ectopic bumps in escapers were reminiscent of mutants in a previously described but molecularly unidentified gene in the region, mal-2, which was recovered in a screen for maternal-effect morphologically abnormal mutants (Hekimi et al. 1995). When we sequenced the Y80D3A.2 coding region of the mal-2 canonical allele qm31, we observed a single-base-pair change in exon 13 coding for a stop within the protein, establishing sel-6 and mal-2 as the same gene. Subsequently, Paula Checchi and William Kelly identified emb-4(hc60) as having a stop mutation in exon 5 of Y80D3A.2, demonstrating that emb-4 also corresponds to this gene (see Checchi and Kelly 2006, accompanying article in this issue).
Alleles of mal-2 and emb-4 were identified in different screens (Miwa et al. 1980; Hekimi et al. 1995), but the three alleles [mal-2(qm31, qm35) and emb-4(hc60)] exhibit completely penetrant maternal-effect embryonic lethality at 25°, as well as maternal-effect embryonic and larval lethality and morphological abnormalities at 20° (Miwa et al. 1980; Hekimi et al. 1995). As three alleles are associated with stop codons in Y80D3A.2 and have completely penetrant and similar phenotypes, we infer that they are all null alleles. We will refer to Y80D3A.2 as emb-4, the first published name of this gene, for the remainder of this article.
The alleles isolated as suppressors of lin-12(n302) are associated with missense mutations and are fully viable. In addition, the suppressor mutation emb-4(sa44) complements the maternal-effect lethality of the null alleles qm31 and hc60 (data not shown; materials and methods). In genetic studies described below, we refer to the viable allele emb-4(sa44) as emb-4(sel) and to the two null alleles emb-4(qm31) and emb-4(hc60) as emb-4(0).
emb-4 interactions with lin-12 and glp-1:
We examined genetic interactions between emb-4 and lin-12 or the lin-12 paralog glp-1, as well as the phenotype of viable emb-4(0) escapers. Our analysis, together with previous studies by Tax et al. (1997), suggests that emb-4 is a positive regulator, but is not absolutely required for lin-12/Notch activity in C. elegans.
The AC/ventral uterine precursor cell decision:
In wild-type hermaphrodites, the gonadal cells Z1.ppp and Z4.aaa interact so that one becomes the AC and the other becomes a ventral uterine (VU) precursor cell (reviewed in Greenwald 2005). In mutants with elevated lin-12 activity, both cells become VUs, whereas loss of lin-12 activity causes both cells to become ACs. When lin-12 activity is elevated, the absence of an AC prevents the formation of a vulva, causing an Egl phenotype that is easy to score in the dissecting microscope. The emb-4(sel) alleles were identified as suppressors of the 0 AC–Egl phenotype (Tax et al. 1997).
The AC/VU decision proceeds normally in emb-4 null mutant animals: 33/33 emb-4(qm31) hermaphrodites raised at 20° have 1 AC, and 15/15 eggs shifted to 25° 1 hr postlaying gave rise to animals that have 1 AC. However, emb-4 appears to be a positive regulator of lin-12 activity in the AC/VU decision, as emb-4(0) alleles can suppress the 0 AC–Egl phenotype associated with lin-12(n302) (Table 1). An emb-4(0) allele does not suppress the 0 AC–Egl phenotype caused by two copies of a stronger lin-12(d) allele, lin-12(n950), but suppresses the 0 AC–Egl phenotype caused by one copy, in lin-12(n950)/lin-12(0) animals (Table 1). In addition, emb-4(sel) enhances the 2AC defect of lin-12(ar170), a hypomorphic allele: 17/70 (25%) lin-12(ar170) hermaphrodites form two anchor cells, but the penetrance of that defect rises to 65% (22/34) in lin-12(ar170); emb-4(sel) animals. These results indicate that suppression is not allele specific, but depends on the level of lin-12 activity.
TABLE 1.
emb-4 suppresses the 0AC-Egl defect caused by lin-12(d) alleles
| Genotype | Egg-laying competent (non-Egl)/total (%) |
|---|---|
| A. emb-4 mutations reduce the activity of lin-12(d) alleles in the AC/VU decision | |
| N2 | >1000 scored (100) |
| lin-12(n302) | >1000 scored (0) |
| lin-12(n302); emb-4(sel) | 88/104 (84) |
| lin-12(n302); emb-4(qm31)a | 6/41 (15) |
| lin-12(n302); emb-4(hc60) | 6/118 (5) |
| lin-12(n950) | >1000 scored (0) |
| lin-12(n950); emb-4(sel) | 92/218 (42) |
| lin-12(n950); emb-4(qm31) | 0/65 (0) |
| lin-12(n950)/lin-12(0)b | 44/143 (31) |
| lin-12(n950)/lin-12(0); emb-4(sel)b | 77/77 (100) |
| lin-12(n950)/lin-12(0); emb-4(qm31)b | 37/63 (59) |
| B. emb-4(0) alleles fail to complement emb-4(sel) for suppression of lin-12(n302) 0AC-Egl defect | |
| lin-12(n302) | >1000 scored (0) |
| lin-12(n302); emb-4(sel)/+ | 1/39 (3) |
| lin-12(n302); emb-4(qm31)/+ | 0/17 (0) |
| lin-12(n302); emb-4(sel)/emb-4(qm31) | 34/42 (81) |
| lin-12(n302); emb-4(hc60)/+ | 0/18 (0) |
| lin-12(n302); emb-4(sel)/emb-4(hc60) | 10/13 (77) |
Of 107 animals, 41 had living progeny; the rest were sterile.
Actual genotype on chromosome III is lin-12(n950)/unc-36(e251) lin-12(n941).
Vulval precursor cell specification:
LIN-12, together with other signaling pathways, specifies the fates of P3.p–P8.p, the six vulval precursor cells (VPCs) (reviewed in Sternberg 2005). In wild-type hermaphrodites, only P5.p and P7.p adopt the vulval 2° fate. Strong hypermorphic lin-12 alleles such as lin-12(n950) cause all six VPCs adopt the 2° fate, resulting in a Muv phenotype. emb-4(sel) and emb-4(0) do not appear to suppress the Muv phenotype caused by lin-12(n950) (Table 2; Tax et al. 1997), but are able to suppress lin-12(n950)/lin-12(0) (Table 2). emb-4(sel) and emb-4(0) alleles are also able to suppress the Muv phenotype associated with arIs53 [lin-12(ΔE)∷gfp], a constitutively active form of LIN-12 (Shaye and Greenwald 2005) (materials and methods; data not shown). These observations suggest that emb-4 also acts as a positive regulator of lin-12 activity in VPC specification.
TABLE 2.
emb-4 affects lin-12 activity in VPC specification
| Average no. of pseudovulvae (n)
|
||
|---|---|---|
| Genotype | non-Egl animals | Egl animals |
| lin-12(n950) | 4.7 ± 0.1 (50) | |
| lin-12(n950); emb-4(sel)a | 4.5 ± 0.2 (28) | |
| lin-12(n950); emb-4(qm31)a | 4.3 ± 0.1 (65) | |
| lin-12(n950)/lin-12(0)b | 2.6 ± 0.1 (44) | 4.2 ± 0.1 (99) |
| lin-12(n950)/lin-12(0); emb-4(sel)b | 0 ± 0.0 (77) | NAc |
| lin-12(n950)/lin-12(0); emb-4(qm31)b | 0.7 ± 0.2 (37) | 2.2 ± 0.2 (26) |
The strains compared were grown and scored in parallel.
Only 0 AC-Egl animals were scored to account only for the role of emb-4 during the VPC specification, and not as a secondary consequence of the AC/VU decision (the presence of an AC decreases the number of pseudovulvae).
Actual genotype on chromosome III was lin-12(n950)/unc-36(e251) lin-12(n941).
All lin-12(n950)/lin-12(0); emb-4(sel) animals were able to lay eggs.
It is notable that the emb-4(0) alleles suppress both the 0 AC–Egl and multivulva defects of lin-12(d) alleles to a lesser degree than the original sel mutations (Tables 1 and 3). The molecular lesions and the fact that emb-4(sel) alleles lack, and complement, the embryonic phenotypes characteristic of emb-4(0) alleles are consistent with these alleles being non-null and affecting only a subset of the activities of the protein. As emb-4(sel) alleles sa44 and n1256 are stronger suppressors than emb-4(0) alleles qm31 and hc60 and are weakly semidominant, they may be dominant negative in action.
TABLE 3.
Cell autonomy of EMB-4 function in the AC/VU decision
The nucleus of Z4 was ablated with a laser microbeam in newly hatched L1 larvae of lin-12(n302) and lin-12(n302); emb-4(sa44) genotypes. Success of ablations was confirmed and the presence of an AC was scored during the late L3 stage.
Number of animals that exhibited the cell fate indicated divided by the total number of animals examined.
Four lin-12(n302) animals were scored in this work; previously, 11 animals were handled identically by Wen et al. (1997).
Embryonic cell fate specification:
Animals that lack zygotic activity of both lin-12 and its paralog, glp-1, arrest development in the L1 stage with a Lag phenotype, one hallmark of which is the absence of a rectum (Lambie and Kimble 1991). All emb-4(0) mutant larvae that we examined have a rectum [122/122 emb-4(hc60) larvae], indicating that, while playing a positive role, emb-4 is not essential for lin-12/Notch activity.
Development of the germline:
Loss of glp-1 activity causes germline precursors to cease mitotic division and to enter meiosis prematurely, resulting in sterility. emb-4(sel) alleles enhance sterility caused by glp-1 loss of function (Tax et al. 1997), suggesting that emb-4 facilitates glp-1 activity in the germline. Constitutive glp-1 activity, as caused by the glp-1(ar202) missense mutation at 25° (Pepper et al. 2003), also causes sterility, caused by excessive proliferation at the expense of meiosis. However, the emb-4(sel) allele sa44 does not suppress the sterility caused by glp-1(ar202): 100% (82/82) of glp-1(ar202); emb-4(sel) hermaphrodites are sterile at 25°, similar to control animals (80/80 sterile). We could not assess suppression of glp-1(ar202) by emb-4(0) because of its lethal null phenotype at 25°.
Cell autonomy of emb-4 functions as a suppressor of lin-12(d):
In wild-type hermaphrodites, when a laser microbeam is used to ablate Z4 (the precursor of Z4.aaa), Z1.ppp always becomes an AC, because the cellular source of the ligand for LIN-12, Z4.aaa, has been removed (Kimble 1981; Seydoux and Greenwald 1989). However, when Z4 is ablated in lin-12(n302) hermaphrodites, Z1.ppp adopts the VU fate, suggesting that LIN-12(n302) is constitutively active in the absence of ligand (Table 3; Greenwald and Seydoux 1990). We have found that when Z4 is ablated in lin-12(n302); emb-4(sel) hermaphrodites, Z1.ppp becomes an AC (Table 3), suggesting that LIN-12(n302) constitutive activity has been lowered. We infer that emb-4 acts cell autonomously to facilitate lin-12 activity in the presumptive VU.
EMB-4 is highly conserved and contains a predicted ATPase domain:
The predicted EMB-4 protein, based on the apparent full-length clone yk1132, contains 1467 amino acids. We analyzed the predicted protein using two programs, SMART (http://smart.embl-heidelberg.de/) (Schultz et al. 1998; Letunic et al. 2006) and BLAST (http://www.ncbi.nih.gov/BLAST/) (Altschul et al. 1997; Schaeffer et al. 2001), and found that EMB-4 shows similarity to ATPases, particularly to helicases of the DEAD-box family (Rocak and Linder 2004). These helicases are characterized by the presence of Walker A- and B-type motifs used for ATP binding, as well as other, family-specific regions of conservation. It is unclear whether EMB-4 is a bona fide member of this family, as it shows homology to only the N-terminal 57% of the consensus DEAD-box helicase domain, including the Walker A motif. The C-terminal part of the ATPase domain shows significant alignment to the consensus sequence of the superfamily I DNA and RNA helicases.
EMB-4 is remarkably conserved in higher eukaryotes, with the BLAST E-value of 0.0 for the human, mouse, fruit fly, and Arabidopsis thaliana orthologs (Figure 1 and see below). There are no other C. elegans proteins highly homologous to it, and other organisms with EMB-4 homologs have only a single copy of the gene encoding this protein. The mouse EMB-4 ortholog, named Aquarius, was recovered in a screen for retinoic-acid-responsive genes (Sam et al. 1998), but functional analysis of Aquarius has not been reported. Schizosaccharomyces pombe has a single EMB-4 ortholog, Cwf11p, which was isolated as a component of a multiprotein complex copurifying with Cdc5p (Ohi et al. 2002). Cdc5p is an essential Myb-related protein implicated in pre-mRNA splicing (Ohi et al. 2002), and the majority of the proteins that copurified with Cdc5p were known pre-mRNA splicing factors. cwf11 also exhibits synergistic genetic interactions with alleles of cdc5 itself (Ohi et al. 2002). We assessed whether feeding animals RNAi constructs targeting the C. elegans orthologs of other Cdc5-complex related proteins (Ohi et al. 2002) could suppress the 0 AC phenotype of lin-12(n302) and lin-12(n379) animals. However, as RNAi for most of these components was highly lethal before the completion of the AC/VU decision (data not shown), we could not determine whether or not the Cdc5p complex proteins positively regulate lin-12 activity in the AC/VU decision.
EMB-4 shows similarity to SMG-2, a helicase required in the process of nonsense-mediated mRNA decay (Figure 1; Page et al. 1999; Grimson et al. 2004). unc-54(r293) mutant animals are uncoordinated and egg laying deficient due to a premature stop codon in the unc-54 mRNA. These phenotypes are suppressible by mutations in all known smg genes (Hodgkin et al. 1989). However, as the loss of emb-4 activity does not suppress either phenotype [120/120 unc-54(r293); emb-4(hc60) animals are Unc-54], there is no evidence for the involvement of EMB-4 in nonsense-mediated decay.
Subcellular localization of EMB-4:
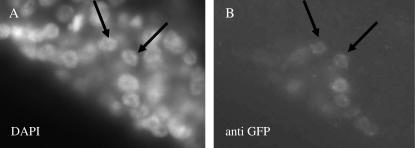
Expression of EMB-4 under the control of a heterologous promoter that is generally expressed appears to be toxic (see materials and methods). Therefore we expressed a GFP-tagged EMB-4 protein under the control of the VPC-expressed lin-31 promoter (Tan et al. 1998) to analyze its subcellular localization. Nuclear staining is observed when transgenic animals are stained with an anti-GFP antibody (Figure 2), consistent with a role for EMB-4 in a nuclear process such as transcription or RNA processing.
Figure 2.—
EMB-4 is localized to the nucleus. Photomicrographs of VPC descendants in an L3 hermaphrodite expressing a GFP∷EMB-4 protein under the control of lin-31 regulatory sequences stained with DAPI (A) or with an antibody against GFP (B). Arrows point to two of the stained vulval nuclei. Anterior is to the left and ventral is down.
DISCUSSION
Genetic analysis demonstrates that emb-4 is a positive regulator of lin-12 activity, as alleles that reduce or eliminate the activity of emb-4 suppress phenotypes associated with constitutively activated forms of LIN-12. Suppression is cell autonomous, consistent with an influence on LIN-12 signal transduction or events that occur downstream of lin-12 signaling. However, viable “escaper” emb-4(0) hermaphrodites do not display the hallmark cell fate transformations associated with loss of lin-12 activity, and arrested larvae do not display defects associated with concomitant loss of lin-12 and glp-1 activity. This observation suggests that emb-4 influences, but is not required for, lin-12/Notch activity in C. elegans or that another gene provides a required but redundant function. However, emb-4(0) mutants arrest with many developmental defects, and the profound phenotypes caused by the absence of emb-4 activity suggest that, in some processes, emb-4 plays a unique or required role. The mechanism(s) by which germ cell precursors are kept transcriptionally quiescent appears to be one such process (Checchi and Kelly 2006, accompanying article in this issue).
EMB-4 has orthologs in fission yeast (but not budding yeast), animals, and plants. The S. pombe ortholog of EMB-4, Cwf11p, copurifies with the Cdc5p complex; many components of this complex are known splicing factors. However, cwf11 differs from most known splicing factors in S. pombe in that it is not an essential gene and therefore was proposed to play only an ancillary role in splicing (Ohi et al. 2002); furthermore, no biochemical evidence for a role in splicing exists at this time. RNAi for most of the C. elegans orthologs of these components was highly lethal, precluding a definitive assessment of their roles in the AC/VU decision. The Senataxin family of proteins, whose ATPase domain is similar to the ATPase domain of EMB-4, has been shown to have both RNA and DNA helicase activities (Kim et al. 1999; Moreira et al. 2004). Whether Cwf11p/EMB-4 has helicase activity is unknown, but the nuclear localization of EMB-4∷GFP suggests a role in a nuclear process.
Recent work from the Kelly lab (H. Furahashi, P. Checchi and W. Kelly, unpublished results) shows that RNAi depletion of ama-1, the gene coding for the large subunit of RNA polymerase II, phenocopies a histone modification defect associated with emb-4(0), suggesting a link between emb-4 and transcription. The link to transcription may be direct, as in participation of EMB-4 in a transcription activation complex or chromatin-modifying complex. Alternatively, it may be indirect, through an effect on RNA metabolism, such as the coupling of transcription with capping, splicing, or 3′-end formation (reviewed in Reed 2003; Zorio and Bentley 2004; Bentley 2005) or in an RNA-dependent chromatin modification process (Bernstein and Allis 2005). A role for EMB-4 in efficient expression of lin-12 itself, a limiting component of the lin-12-signaling system, or key target genes, by any of the above mechanisms, could account for its function as a positive regulator of lin-12 activity.
Acknowledgments
We gratefully acknowledge Paula Checchi and William Kelly for many fruitful discussions, comments on the manuscript, and sharing unpublished data. We thank Richard Ruiz and Xinlan Zhou for excellent technical assistance, Natalie de Souza for comments on the manuscript, and past and present members of the Greenwald laboratory for useful ideas. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources. This work was supported by NIH grant CA095389 (to I.G.). I.K. is currently a Postdoctoral Associate, and I.G. is an Investigator, of the Howard Hughes Medical Institute.
References
- Altschul, S. F., T. L. Madden, A. A. Schaeffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, D. L., 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17: 251–256. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., and C. D. Allis, 2005. RNA meets chromatin. Genes Dev. 19: 1635–1655. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada, R., E. Isnenghi, M. Culotti and G. von Ehrenstein, 1981. Genetic analysis of temperature-sensitive embryogenesis mutants in Caenorhabditis elegans. Dev. Biol. 84: 193–205. [DOI] [PubMed] [Google Scholar]
- Checchi, P. M., and W. G. Kelly, 2006. emb-4 is a conserved gene required for efficient germline-specific chromatin remodeling during Caenorhabditis elegans embryogenesis. Genetics 174: 1895–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald, I., 2005. LIN-12/Notch signaling in C. elegans, in WormBook, edited by The C. elegans Research Community (doi/10.1895/wormbook.1891.1810.1891; http://www.wormbook.org).
- Greenwald, I., and G. Seydoux, 1990. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature 346: 197–199. [DOI] [PubMed] [Google Scholar]
- Greenwald, I. S., P. W. Sternberg and H. R. Horvitz, 1983. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34: 435–444. [DOI] [PubMed] [Google Scholar]
- Grimson, A., S. O'Connor, C. L. Newman and P. Anderson, 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol. Cell. Biol. 24: 7483–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekimi, S., P. Boutis and B. Lakowski, 1995. Viable maternal-effect mutations that affect the development of the nematode Caenorhabditis elegans. Genetics 141: 1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert, O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., A. Papp, R. Pulak, V. Ambros and P. Anderson, 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, E. J., Q. Dong and I. Greenwald, 1996. Evidence for physical and functional association between EMB-5 and LIN-12 in Caenorhabditis elegans. Science 273: 112–115. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Karp, X., and I. Greenwald, 2003. Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 17: 3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. D., J. Choe and Y. S. Seo, 1999. The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry 38: 14697–14710. [DOI] [PubMed] [Google Scholar]
- Kimble, J., 1981. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev. Biol. 87: 286–300. [DOI] [PubMed] [Google Scholar]
- Lambie, E. J., and J. Kimble, 1991. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development 112: 231–240. [DOI] [PubMed] [Google Scholar]
- Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz et al., 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34: D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa, J., E. Schierenberg, S. Miwa and G. von Ehrenstein, 1980. Genetics and mode of expression of temperature-sensitive mutations arresting embryonic development in Caenorhabditis elegans. Dev. Biol. 76: 160–174. [DOI] [PubMed] [Google Scholar]
- Moreira, M. C., S. Klur, M. Watanabe, A. H. Nemeth, I. Le Ber et al., 2004. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 36: 225–227. [DOI] [PubMed] [Google Scholar]
- Ohi, M. D., A. J. Link, L. Ren, J. L. Jennings, W. H. McDonald et al., 2002. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22: 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, M. F., B. Carr, K. R. Anders, A. Grimson and P. Anderson, 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19: 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper, A. S., D. J. Killian and E. J. Hubbard, 2003. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163: 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess, J. R., and D. I. Hirsh, 1986. Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev. Biol. 117: 156–173. [DOI] [PubMed] [Google Scholar]
- Reed, R., 2003. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15: 326–331. [DOI] [PubMed] [Google Scholar]
- Rocak, S., and P. Linder, 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5: 232–241. [DOI] [PubMed] [Google Scholar]
- Sam, M., W. Wurst, M. Kluppel, O. Jin, H. Heng et al., 1998. Aquarius, a novel gene isolated by gene trapping with an RNA-dependent RNA polymerase motif. Dev. Dyn. 212: 304–317. [DOI] [PubMed] [Google Scholar]
- Schaeffer, A. A., L. Aravind, T. L. Madden, S. Shavirin, J. L. Spouge et al., 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29: 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierenberg, E., J. Miwa and G. von Ehrenstein, 1980. Cell lineages and developmental defects of temperature-sensitive embryonic arrest mutants in Caenorhabditis elegans. Dev. Biol. 76: 141–159. [DOI] [PubMed] [Google Scholar]
- Schultz, J., F. Milpetz, P. V. Bork and C. P. Ponting, 1998. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 95: 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux, G., and I. Greenwald, 1989. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell 57: 1237–1245. [DOI] [PubMed] [Google Scholar]
- Shaye, D. D., and I. Greenwald, 2005. LIN-12/Notch trafficking and regulation of DSL ligand activity during vulval induction in Caenorhabditis elegans. Development 132: 5081–5092. [DOI] [PubMed] [Google Scholar]
- Sternberg, P. W., 2005. Vulval development, in WormBook, edited by The C. elegans Research Community (doi/10.1895/wormbook.1891.1896.1891; http://wwww.wormbook.org).
- Tan, P. B., M. R. Lackner and S. K. Kim, 1998. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell 93: 569–580. [DOI] [PubMed] [Google Scholar]
- Tax, F. E., J. H. Thomas, E. L. Ferguson and H. R. Horvitz, 1997. Identification and characterization of genes that interact with lin-12 in Caenorhabditis elegans. Genetics 147: 1675–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., and A. Fire, 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Timmons, L., D. L. Court and A. Fire, 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- Wen, C., M. M. Metzstein and I. Greenwald, 1997. SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN-12/NOTCH signalling. Development 124: 4759–4767. [DOI] [PubMed] [Google Scholar]
- Zorio, D. A., and D. L. Bentley, 2004. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 296: 91–97. [DOI] [PubMed] [Google Scholar]