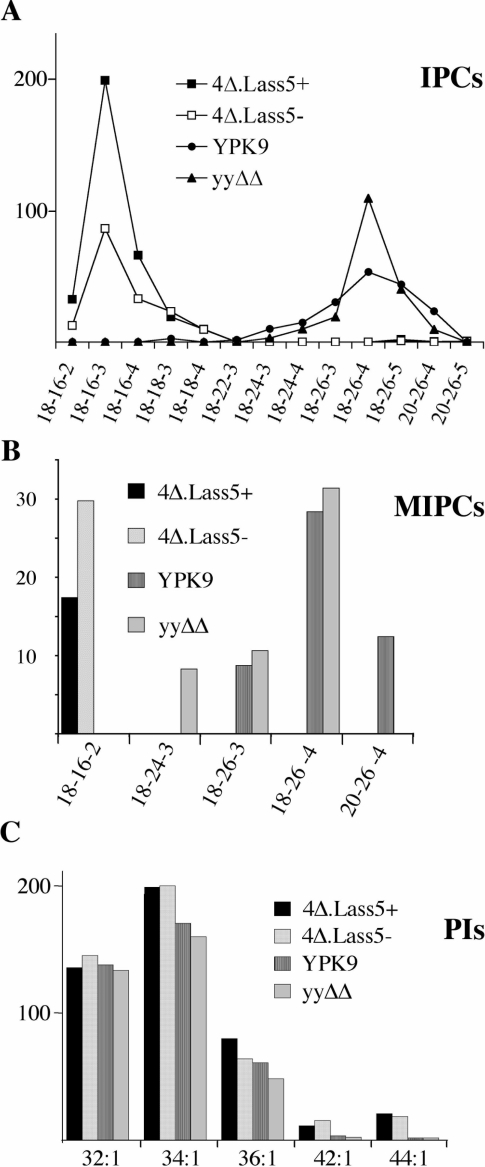
Figure 4. Sphingolipids from 4Δ.Lass5 cells contain mainly C16:0 and C18:0 fatty acids by HPLC–ESI-MS/MS.
All sphingolipid classes were extracted quantitatively [35] from cells growing exponentially in LM at 30 °C and analysed by HPLC–ESI-MS/MS. YPK9, ypc1Δ ydc1Δ (yyΔΔ) and 4Δ.Lass5 cells were all grown with methionine (4Δ.Lass5+), 4Δ.Lass5 also without methionine (4Δ.Lass5−). Parent ion and fragment ion masses (m/z) were recorded throughout the HPLC run and were screened for all theoretically possible sphingolipids containing different saturated, monounsaturated, mono- or di-hydroxylated fatty acids combined with DHS or PHS as well as for conventional phospholipids [40]. The vertical axis in all panels gives the ion counts in thousands. All species shown in (A) were identified as phosphoinositides by characteristic fragment ions at m/z 259 and 241, corresponding to inositolphosphate and inositolphosphate–H2O, except for those that were less than 1% of the most predominant IPC. However, the LCB could not be identified by fragmentation and sphingolipids were annotated by three consecutive numbers to indicate the probable number of carbon atoms in the LCB, the probable number of carbon atoms in the fatty acid, and the sum of hydroxy groups in the ceramide moiety respectively. (A) The lipid extracts were mixed with lipid extracts from WT cells grown in [13C]glucose as the only carbon source and the results were normalized using this internal standard. The normalized number of ions derived from IPCs present in extracts of 4Δ.Lass5+, 4Δ.Lass5− and ypc1Δ ydc1Δ amounted to 183%, 92% and 106% of the WT (YPK9) respectively. (C) The PIs are specified by two consecutive numbers indicating the total number of carbon atoms and double bonds present in the two fatty acids respectively. The Figure shows one representative of several experiments yielding very similar results.