Abstract
LINE-1s (long interspersed nuclear elements-1) are abundant non-LTR retrotransposons that comprise 17% of the human genome. The 5′ untranslated region (5′UTR) of human L1 (L1Hs) houses a poorly understood internal promoter. Here we report that mutations at a putative runt-domain transcription factor (RUNX) site (+83 to +101) in the 5′UTR decreased L1Hs transcription and retrotransposition in cell culture-based assays. Exogenous expression of RUNX3, but not the other two RUNX family members, RUNX1 and RUNX2, increased L1Hs transcription and retrotransposition, which were otherwise decreased by siRNAs targeting RUNX3 and a dominant negative RUNX. Further more, the specific interaction between RUNX3 and its binding site was demonstrated by an electrophoretic mobility shift assay (EMSA) using an anti-RUNX3 antibody. Interestingly, RUNX3 may also regulate the antisense promoter activity of L1Hs 5′UTR via another putative RUNX site (+526 to +508), as revealed by site-directed mutations and exogenous expression of RUNX factors. Our results indicate an important role for RUNX3 in L1Hs retrotransposition as well as transcription from its 5′UTR in both sense and antisense directions, and they should contribute to our understanding of the mechanism underlying L1Hs retrotransposition and its impact on the expression of adjacent cellular genes.
INTRODUCTION
LINE-1s (long interspersed nuclear elements-1) or L1s are highly abundant retrotransposons comprising 17% of the human genome (1,2). Although most human L1s (L1Hs) are retrotransposition-defective due to 5′ truncation, internal rearrangement or mutation (3), 80–100 L1Hs are potentially retrotransposition-competent in the diploid genome (4). A full-length L1Hs is ∼6 kb and contains a 5′ untranslated region (5′UTR), two non-overlapping open reading frames (ORF1 and ORF2) and a 3′UTR (5,6). The original retrotransposon is transcribed into RNA and exported to the cytoplasm, where ORF1 and ORF2 proteins are translated and a ribonucleoprotein particle that includes L1 RNA, ORF1p and possibly ORF2p is formed (7,8). ORF1 encodes a 40 kDa protein with RNA binding activity (7,9); ORF2 encodes a protein with endonuclease (10) and reverse transcriptase (RT) (11) activities. Both ORF1p and ORF2p are required for retrotransposition (10,12). L1s integrate into the genome by target primed reverse transcription using the free 3′-OH at the endonuclease cut site as a primer and the L1 RNA as a template (13). The integrated L1 is generally flanked by perfect 7–20 bp target site duplications. L1 retrotransposons continuously shape the human genome by insertion, non-homologous recombination, transduction of 3′ and 5′ flanking sequences (14,15) and mobilization of other transposable elements, such as Alu elements (16), processed pseudogenes (17) and SVA elements (5).
The 5′UTR of L1Hs houses an internal promoter (18,19). Transcription of L1Hs is initiated at the +1 nt and is proposed as the rate-limiting step in the process of L1Hs retrotransposition (18–20). Deletion analysis revealed that the first 100 (18) or 155 bp (19) contains cis-elements critical for L1Hs transcription, which are likely regulated by transcription factors because the 5′UTR contains binding sites for certain cellular proteins (19,21). For example, a perfect Yin Yang-1 (YY-1) core binding sequence lies between +13 and +21, and several groups have demonstrated binding of YY-1 to this region (19,22). YY-1 may serve to position the transcription initiation complex rather than to regulate the transcription level (23). Also, SOX family members (24) and steroid hormones (25) have been found to influence L1Hs promoter activity. Finally, L1Hs transcription and retrotransposition are repressed by methyl-CpG-binding protein 2 (26). Most interestingly, an antisense promoter (ASP) has been identified at +400 to +600 that drives transcription of adjacent cellular genes (27,28). However, the regulation of L1Hs transcription, especially its relationship with retrotransposition, is still poorly understood.
The RUNX (runt-domain transcription factor) family contains heterodimeric transcription factors composed of α and β subunits (29). The α subunit is a homolog of the product of the Drosophila melanogaster segmentation gene, runt, and contains a conserved 128-amino acid region, the runt homology domain, which is required for DNA binding and heterodimerization with the β subunit. The β subunit fails to bind DNA on its own but increases the affinity of the RUNX factors for DNA binding. Only one mammalian gene encoding the β subunit is known; however, three mammalian α subunit genes have been identified: RUNX1//PEBP2αB/CBFA2/AML1, RUNX2/PEBP2αA/CBFA1/AML3 and RUNX3/PEBP2αC/CBFA3/AML2 (30). Following the nomenclature recommendation, RUNX terminology is used here. The RUNX factors act on tissue-specific genes whose promoter or enhancer contains the consensus runt-domain core binding sequence TGT/CGGT (31). RUNX1 is essential for definitive hematopoiesis (32–34). RUNX2 is essential for osteogenesis (35). RUNX3 regulates growth of gastric epithelial cells (36), development and survival of TrkC dorsal root ganglia (DRG) neurons (37), as well as axonal projection and path finding of subtypes of DRG neurons (38,39); it is also required for establishment of epigenetic silencing in cytotoxic lineage thymocytes (40) and T cell development during thymopoiesis (41).
Here we show that mutations of a putative RUNX site dramatically decrease L1Hs transcription and retrotransposition. Exogenous expression of RUNX3, but not RUNX1 or RUNX2, significantly increases L1Hs transcription and retrotransposition, which are otherwise decreased by a dominant negative form of RUNX or siRNAs targeting RUNX3. In addition, we find that RUNX3 may also regulate the ASP activity of L1Hs 5′UTR. Our study therefore indicates an important role for RUNX3 in L1Hs transcription and retrotransposition.
MATERIALS AND METHODS
Cell culture
HeLa and human osteosarcoma cell line 143B (ATCC, Manassas, VA) were grown in high glucose Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin and 0.1 mg/ml streptomycin (all from Invitrogen, Carlsbad, CA) in a humidified, 5% CO2 incubator at 37°C.
Plasmid constructs
Mammalian expression plasmids containing the cDNA of human RUNX1 (pEF-Bos-PEBP2αB1), RUNX2 (pEF-Bos-PEBP2αA), RUNX3 (pEF-Bos-PEBP2αC1), the fusion gene of AML1/RUNX1 and Eight-Twenty One oncoprotein (ETO) (pEF-Bos-AML1/ETO) and the control vector pEF-Bos were generously provided by Dr Yoshiaki Ito (Institute of Molecular and Cell Biology, Singapore) (42).
To construct luciferase reporter plasmids, 5′UTRs of the wild-type L1RP, L1.3 or the mutated L1 elements were amplified using Expand High Fidelity PCR System (Roche, Mannheim, Germany) and inserted upstream of the luciferase gene in pGL3-Basic vector (Promega, Madison, WI), in either sense or antisense direction. Sequences of L1 5′UTR in all these reporter constructs were verified by DNA sequencing (DNA Sequencing Core, University of Pennsylvania).
To perform vector-based RNAi, DNA oligonucleotides targeting RUNX3 at different locations were designed with siRNA Target Finder and Design Tool at www.ambion.com/techlib/misc/siRNAfinder.html (Ambion, Austin, TX), and inserted into ApaI–EcoRI linearized pSilencer 1.0 vector (Ambion) according to the manufacturer’ instructions. Control RNAi plasmid was constructed by insertion of a sequence with limited sequence identity to any known sequences in the human genome. All the inserted sequences were verified by sequencing.
Site-directed mutagenesis
Site-directed mutagenesis was performed using QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following the manufacturer’s instructions. The introduced mutations were verified by sequencing.
Random mutagenesis
Random mutagenesis of L1Hs 5′UTR was performed using GeneMorph PCR Mutagenesis Kit (Stratagene) according to the manufacturer’s protocols. The mutation frequency was controlled at one to two mutations per 500 bp. The mutagenic PCR product was purified with MinElute PCR Purification Kit (Qiagen, Valencia, CA) and swapped into the random mutagenesis backbone. Hundreds of colonies containing L1s with random mutation(s) were obtained and the mutations were identified by sequencing.
Real-time quantitative RT–PCR and nested RT–PCR
Total RNA was isolated from 1 × 106 cultured 143B or HeLa cells with TRIzol reagent (Invitrogen). After treatment with RNase-free DNase (Invitrogen), RNA was further purified with RNeasy RNA Isolation Kit (Qiagen) and reverse-transcribed using Superscript First-Strand Synthesis Kit for RT–PCR (Invitrogen) under conditions described by the suppliers. Real-time quantitative PCR using cDNA synthesized above or from the Human Multiple Tissue cDNA Panels (Clontech, Palo Alto, CA) was performed with Sybergreen PCR Core Reagent Kit (Applied Biosystems, Foster City, CA) on the ABI Prism 7700 Sequence Detection System (Applied Biosystems) according to the manufacturers’ instructions. Real-time RT–PCR of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as an internal control. To avoid false positive results due to amplification of contaminating genomic DNA in the cDNA preparation, each pair of primers was designed to reside in exons separated by an intron. PCR products were detected as a single band on gel electrophoresis.
RT–PCR was performed to detect full-length RUNX3 mRNA in 143B and HeLa cells with primer pairs covering the N- and C-terminal regions of RUNX3, respectively, as described previously (36). In particular, a nested PCR was performed to amplify the N-terminal region of RUNX3, in which first round PCR was carried out in a 25 µl system using PCR Core Kit (Roche) with primers 5′-CGCCACTTGATTCTGGAGGATTTGT-3′ and 5′-TGAAGTGGCTTGTGGTGCTGAGTGA-3′, and 3 µl of product was used for the second round PCR with primers 5′-TATTCCCGTAGACCCAAGCA-3′ and 5′-GTCTGGTCCTCCAGCTTCTG-3′. Primers for the C-terminal region are: 5′-GAGTTTCACCCTGACCATCACTGTG-3′ and 5′-GCCCATCACTGGTCTTGAAGGTTGT-3′.
Luciferase reporter assay
143B or HeLa cells were seeded in 6-well plates at 3 × 105 cells/well and grown overnight to ∼40% confluence prior to transfection. To test the promoter activity of different L1 5′UTRs, a total of 0.5 µg sense or antisense reporter construct and 0.01 µg pRL-TK internal control (Promega) were used for each transfection. All transfection experiments were done in triplicate and repeated at least twice with different DNA isolates. Forty-eight hours post-transfection, luciferase analysis was performed on Luminoskan Ascent (Thermo-Labsystems, Franklin, MA) using Dual-Luciferase Reporter Assay System (Promega) according to the manufacturers’ instructions. For co-transfection experiments, 1 µg of each RUNX or control expression plasmid was used.
Retrotransposition assay
The retrotransposition assay was performed as described previously (43). Each transfection received 1 µg plasmid containing L1 element tagged with the EGFP cassette alone or in a co-transfection with 1 µg expression plasmid of a RUNX factor. For each assay, a negative control of pL1RP(JM111)-EGFP(puro), which contains an inactive L1 element lacking retrotransposition activity due to two missense mutations in ORF1 and a positive control of pL1.3-EGFP(puro) was performed. Antibiotic selection with puromycin (10 µg/ml) was started 48 h post-transfection. On day 6 post-transfection, fluorescence-activated cell scanning (FACS) was performed and cells were counted as positive when they showed greater fluorescence intensity than the most fluorescent cell transfected with pL1RP(JM111)-EGFP(puro).
RNAi
For RNAi experiments using the luciferase reporter assay, cells were transfected as described above except that 1 µg of each vector-based RNAi construct targeting RUNX3 or the control plasmid and 0.5 µg of pCEP4-puro were transfected in each well together with 0.25 µg L1 5′UTR reporter construct and 0.01 µg pRL-TK internal control. Forty-eight hours post-transfection, cells were changed to complete medium containing 1 µg/ml puromycin. Puromycin selection continued until the cells were lysed for the luciferase assay on day 5 post-transfection. For RNAi experiments with the retrotransposition assay, all procedures were as described above except that 1 µg of each RNAi construct was co-transfected with 1 µg pL1RP-EGFP(puro).
Electrophoretic mobility shift assay (EMSA) and supershift EMSA
Nuclear extract from 143B cells was prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents plus Halt Protease Inhibitor Cocktail Kit (Pierce, Rockford, IL) following the manufacturer’s protocols and stored at –80°C until used. 5′-Biotin-labeled DNA oligos containing the wild-type (5′-CTGCATTTCCATCTGAGGTAC-3′) or mutated RUNX site (5′-CTGCATTTCCATCTGAGGCAC-3′) were synthesized and the two complementary oligos were annealed to obtain the double-stranded probe. EMSA and supershift EMSA were performed using LightShift Chemiluminescent EMSA Kit (Pierce). Five micrograms of nuclear extract and 30 fmol of labeled probe alone or with 9 pmol unlabeled probe were incubated in the 10× binding buffer plus 1 µg poly(dI–dC) in 20 µl reaction system at room temperature for 20 min. For supershift EMSA, 1 µl rabbit anti-human RUNX3 antibody (a generous gift from Dr Yoram Groner, Weizmann Institute of Science, Israel) (44) or normal rabbit IgG was incubated with 5 µg of nuclear extract on ice for 30 min in buffer containing 2 µg poly(dI–dC), 0.68 mM EDTA, 6.8 mM HEPES and 1.36% Ficoll 400 before addition of 30 fmol labeled probe and further incubation on ice for 30 min as described previously (45). Both incubation conditions worked for EMSA and supershift EMSA; however, only the condition with optimal sensitivity was used in formal experiments. The entire 20 µl binding reaction was loaded on a 7.5% polyacrylamide gel and run at room temperature in 0.25× TBE at 110 V for 1–1.5 h. The electrophoresed binding reaction was then transferred to BrightStar-Plus positively charged Nylon membrane (Ambion) in Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad, Hercules, CA) at 15 V for 30 min and cross-linked in UV Stratalinker 1800 (Stratagene) at auto cross-link level for 1 min. Detection of Biotin-labeled DNA probe was performed strictly following the manufacturer’s protocol.
Bioinformatics
A search for potential transcription factor binding sites on the L1RP 5′UTR was performed using the program Mat Inspector V2.2 at www.genomatix.de/cgi-bin/matinspector/matinspector.pl (46). Comparison of the 5′UTRs of full-length L1Hs was executed using ClustalW (47).
Statistical analysis
Statistical analysis of data was performed using the SPSS statistics software package (SPSS, IL). All results were expressed as mean ± SD, and P < 0.05 was used for significance.
RESULTS
A single-nucleotide mutation in the 5′UTR significantly inhibits L1Hs retrotransposition
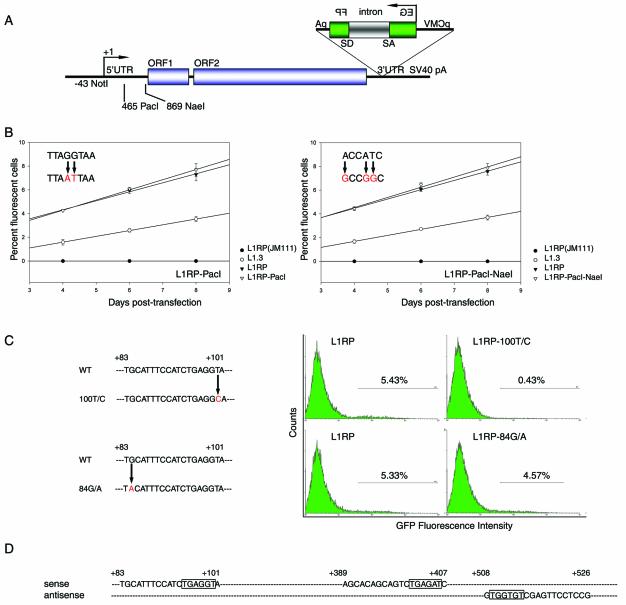
To study the regulation of L1Hs retrotransposition, we used random mutagenesis to identify important sequences in L1RP, one of the most active known L1Hs (43,48). To construct the backbone for random mutagenesis of the first half of 5′UTR, a PacI site was created at +465 to +472 of L1RP to produce pL1RP-PacI-EGFP(puro) (Fig. 1A). Because undesired mutation(s) might also be introduced during the mutagenic process and exert a functional effect, retrotransposition activities of different clones of pL1RP-PacI-EGFP(puro) were tested and compared with that of pL1RP-EGFP(puro). The assay to test retrotransposition activity is based on an EGFP cassette inserted in the antisense direction in the 3′UTR of L1 element (43). The EGFP gene is driven by the CMV immediate early promoter, followed by the TK poly(A) signal. The EGFP gene is disrupted by a γ-globin intron oriented in the sense direction relative to L1 such that it can only be spliced out of L1 RNA but not EGFP RNA. Therefore, only retrotransposition of the tagged L1 element can generate a functional EGFP gene. EGFP-positive cells are then detected by FACS and the percent of fluorescent cells represents the retrotransposition activity of the tagged L1 element. In this study, the pL1RP-PacI-EGFP(puro) clone with the closest retrotransposition activity to pL1RP-EGFP(puro) was used as the backbone for random mutagenesis (Fig. 1B, left). Similarly, we constructed the backbone for random mutagenesis of the second half of L1RP 5′UTR by creating an NaeI site at +869 to +874 (Fig. 1A and B, right). To facilitate data interpretation, the random mutation frequency was controlled at one to two mutations per 500 bp. Sequencing revealed that most L1 elements obtained from random mutagenesis contained 1–2 nt substitutions.
Figure 1.
Potentially important RUNX sites in the 5′UTR for L1Hs retrotransposition. (A) Construction of backbones for random mutagenesis of L1RP 5′UTR. L1Hs contains 5′ and 3′UTRs and two ORFs. Unique PacI and NaeI sites were created at +465 to +472 and +869 to +874 in the L1RP 5′UTR, respectively. An EGFP cassette to detect L1Hs retrotransposition is inserted within the 3′UTR in the antisense direction and consists of the CMV immediate early promoter (pCMV), the EGFP gene disrupted by a sense direction oriented γ-globin intron (intron) and the TK poly(A) signal (pA). (B) The created PacI (left) and NaeI (right) sites have no effect on retrotransposition. Retrotransposition activities of the random mutagenesis backbones for the 5′UTR, i.e. L1RP-PacI and L1RP-PacI–NaeI (empty triangle), the wild-type L1RP (solid triangle), a negative control of L1RP(JM111) (solid circle) and a positive control of L1.3 (empty circle) were tested by FACS on days 4, 6 and 8 post-transfection (n = 6). The original sequence and created mutations are shown in the upper left region of the corresponding retrotransposition activity curve. (C) Two single-nucleotide mutations, 100T/C and 84G/A, identified by random mutagenesis at a potential RUNX site and their retrotransposition activities in 143B cells (n = 6). One representative FACS result of each sample is shown. (D) Three potential RUNX sites in the L1RP 5′UTR. Each site consists of 19 nt, and the consensus core RUNX sequences are shown (boxed). The first two RUNX sites are on the sense strand, and the third on the antisense strand.
Among the 50 5′UTR-mutated L1 elements tested to date, we found a single-nucleotide mutation (100T/C) that significantly reduced retrotransposition activity to 8% of wild-type L1RP (P < 0.001, Fig. 1C, upper).
Mutations at the first but not the second or third RUNX site inhibit L1Hs retrotransposition
Since this single-nucleotide mutation so greatly reduced retrotransposition activity, we hypothesized that it may reside in a critical cis-element recognized by a transcription factor. Therefore, we explored potential transcription factor binding sites in L1RP 5′UTR in silico and obtained 94 matches. The 100T/C mutation fell within a potential binding site (+83 to +101) for RUNX factors (Fig. 1D). In addition, two other RUNX sites were identified in the L1RP 5′UTR, one at +389 to +407 and the other at +526 to +508. Interestingly, the first two RUNX sites were on the sense strand, whereas the third was on the antisense strand. We further aligned the 5′UTRs of 20 L1Hs with known retrotransposition activity. The three RUNX sites were completely conserved in all 20 elements (data not shown). When we re-examined the remaining randomly mutated L1 clones, a second clone was found to contain a single-nucleotide mutation (84G/A) residing within the first RUNX site. This mutation decreased retrotransposition activity to 85% of wild-type L1RP (P = 0.029, Fig. 1C, bottom).
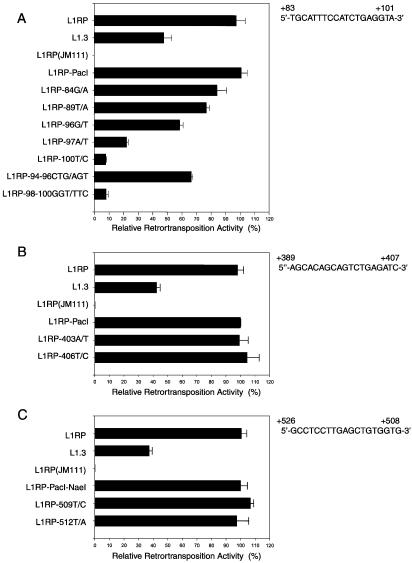
To further study the potential RUNX sites, we created additional nucleotide mutations at those sites. Altogether, single-nucleotide mutations, 89T/A, 96G/T and 97A/T, as well as 3-nt mutations, 94–96CTG/AGT and 98–100GGT/TTC, were made at the first RUNX site; 403A/T and 406T/C mutations were made at the second site; and 509T/C and 512T/A mutations were made at the third site. We found that all mutations at the first RUNX site reduced L1Hs retrotransposition activity (Fig. 2A). However, mutations at the second and third RUNX sites had no significant effect (Fig. 2B and C), which was not very surprising since both sites were outside the first 100 bp of 5′UTR and the core sequence of the second site differed from the consensus sequence by 1 nt.
Figure 2.
Mutations at the first but not the second or third RUNX site inhibit L1Hs retrotransposition in 143B cells. (A) Retrotransposition activities of the internal controls, i.e. L1RP, L1.3 and L1RP(JM111), and L1Hs containing mutations at the first RUNX site, 84G/A, 89T/A, 96G/T, 97A/T, 100T/C, 94–96CTG/AGT and 98–100GGT/TTC, as normalized to that of L1RP-PacI. (B) Retrotransposition activities of the L1Hs elements containing mutations, 403A/T and 406T/C, at the second RUNX site, as normalized to that of L1RP-PacI. (C) Retrotransposition activities of the L1Hs elements containing mutations, 509T/C and 512T/A, at the third RUNX site, as normalized to that of L1RP-PacI–NaeI (n = 6).
The first RUNX site in the 5′UTR is critical for L1Hs transcription
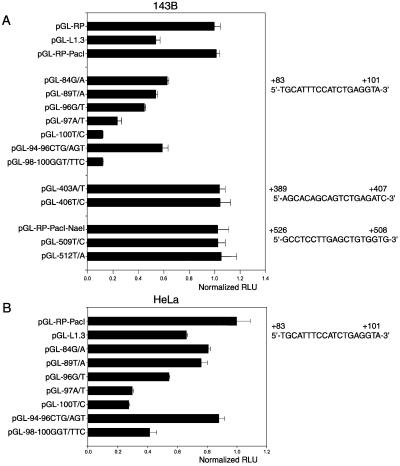
Because members of the RUNX family are transcription regulators and transcription has been proposed as the rate-limiting step in L1Hs retrotransposition (19,20), we examined the promoter activities of wild-type and mutated L1Hs 5′UTRs by luciferase reporter assays. We found that the promoter activities of the mutated L1Hs elements agreed well with their corresponding retrotransposition activities, i.e. mutations at the first RUNX site reduced the promoter activity of the L1Hs 5′UTR whereas mutations at the second and third RUNX sites had no effect (Fig. 3A). Similar results were observed in HeLa cells (Fig. 3B) except that promoter activities of all the tested 5′UTRs were lower than their counterparts tested in 143B cells.
Figure 3.
Mutations at the first RUNX site decrease L1Hs transcription. (A) Promoter activities of the wild-type and mutant L1Hs 5′UTRs in 143B cells are normalized to that of pGL-RP-PacI (RLU, relative luciferase units). Wild-type sequences for the three RUNX sites are shown. (B) Promoter activities of the wild-type and mutant L1Hs 5′UTRs in HeLa cells (n = 9).
Exogenous expression of RUNX3 increases L1Hs transcription and retrotransposition
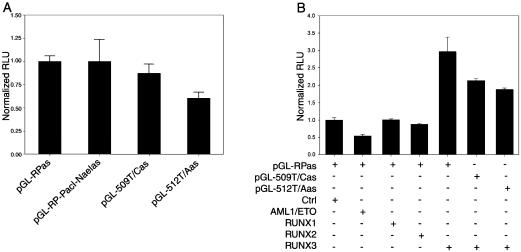
To provide additional evidence that the RUNX factor(s) indeed regulated L1Hs transcription and retrotransposition and to identify the responsible RUNX factor(s), eukaryotic expression plasmids containing RUNX factors were co-transfected into 143B cells in both the luciferase and retrotransposition assays. We found that the dominant negative form of RUNX (dn-RUNX), a fusion protein of RUNX1 and ETO from a translocation, t(8:21)(22q;22q) (49,50), significantly decreased the promoter activity of L1RP 5′UTR by 56% (P < 0.001, Fig. 4A). Although following transfection all three RUNX factors were expressed at comparable levels as revealed by real-time quantitative RT–PCR (data not shown), only RUNX3 significantly increased the promoter activity of L1RP 5′UTR by 2.1-fold (P < 0.001); and this effect could be almost completely abolished by the 100T/C mutation at the first RUNX site. To investigate the effects of RUNX factors in vivo, the L1Hs retrotransposition activity was tested in the presence of exogenous expression of different RUNX factors. In agreement with the results of the luciferase reporter assay, dn-RUNX significantly decreased L1 retrotransposition by 70% (P < 0.001, Fig. 4B). RUNX3 increased the L1Hs retrotransposition activity by 15%. However, L1Hs retrotransposition remained nearly abolished by the 100T/C mutation in the presence of RUNX3, suggesting the specific interaction between RUNX3 and this site. Surprisingly, RUNX1 and RUNX2 decreased L1Hs retrotransposition activity. When we repeated this retrotransposition experiment in HeLa cells, we obtained very similar results (Fig. 4C). However, the effects of RUNX3 and dn-RUNX were greatly magnified; RUNX3 increased retrotransposition activity by 70% (P = 0.025) and dn-RUNX decreased it by 87% (P = 0.010). On the other hand, RUNX1 and RUNX2 had no significant effect.
Figure 4.
Exogenous expression of RUNX3 increases L1Hs transcription and retrotransposition. (A) In 143B cells, exogenous expression of RUNX3, but not RUNX1 or RUNX2, significantly increases the promoter activity of L1Hs 5′UTR compared with the empty vector control (Ctrl), and this effect of RUNX3 is abolished by mutation (100T/C). In addition, dn-RUNX (AML1/ETO) significantly decreases the promoter activity of 5′UTR (n = 9). (B) In 143B cells, exogenous expression of RUNX3 increases the L1Hs retrotransposition activity by 15%, and this effect of RUNX3 is abolished by mutation (100T/C). dn-RUNX dramatically decreases the L1Hs retrotransposition activity. RUNX1 and RUNX2 also decrease the L1Hs retrotransposition activity (n = 6). (C) In HeLa cells, retrotransposition activity of L1Hs is significantly increased upon exogenous expression of RUNX3 and decreased by dn-RUNX. RUNX1 and RUNX2 have no effect on L1Hs retrotransposition (n = 6).
RNAi of RUNX3 decreases L1Hs transcription and retrotransposition
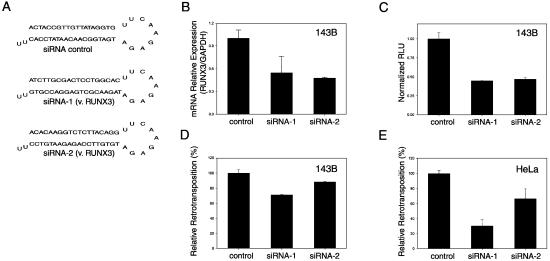
To further confirm that RUNX3 did play an important role in L1Hs transcription and retrotransposition, we employed vector-based RNAi of RUNX3 using two siRNA sequences targeting different locations of RUNX3 (Fig. 5A). In 143B cells, both RNAi constructs decreased RUNX3 mRNA level by ∼50% as revealed by real-time quantitative RT–PCR (P = 0.035 and <0.001, respectively, Fig. 5B). Co-transfection using either RNAi construct decreased the promoter activity of L1Hs 5′UTR by 56 and 54% (P = 0.007 and 0.005, respectively, Fig. 5C) and the L1Hs retrotransposition by 29 and 12% (P = 0.007 and 0.041, respectively, Fig. 5D) compared with the control (Fig. 5A). Again, RNAi of RUNX3 in HeLa cells had a significantly greater effect, decreasing L1Hs retrotransposition by up to 70% (P = 0.002 and 0.040, respectively, Fig. 5E).
Figure 5.
RNAi of RUNX3 decreases L1Hs transcription and retrotransposition. (A) Schematic figure of the hairpin siRNA products from the vector-based RNAi constructs for control (siRNA control) and RUNX3-targeting (siRNA-1 and siRNAi-2 versus RUNX3). (B) Real-time quantitative RT–PCR reveals that the RUNX3 mRNA level is reduced in 143B cells transfected with either RNAi construct targeting RUNX3 (siRNAi-1 and siRNAi-2) compared with the control. (C) In 143B cells, both RNAi constructs decrease the promoter activity of L1Hs 5′UTR. (D) In 143B cells, both RNAi constructs decrease the L1Hs retrotransposition activity. (E) In HeLa cells, both RNAi constructs decrease the L1Hs retrotransposition activity by 33 and 70%, respectively (n = 6).
Mutation at the first RUNX site impairs its interaction with RUNX3
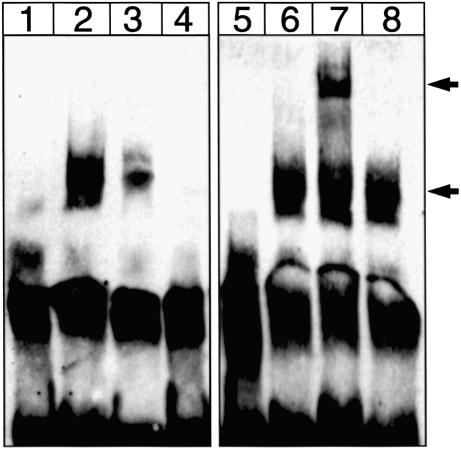
To further investigate whether mutation at the first RUNX site inhibited its interaction with protein, we performed EMSA using 143B nuclear extract and probes containing either the wild-type or the 100T/C mutant first RUNX site. DNA–protein interaction was significantly reduced when the wild-type probe (Fig. 6, lane 2) was replaced with the mutant probe (lane 3). We then carried out a super-shift EMSA using rabbit anti-human RUNX3 antibody, and thereby demonstrated a specific interaction of RUNX3 with this site. A super-shifted band was observed when anti-RUNX3 antibody was present (lane 7), but this band was absent when normal rabbit IgG was used (lane 8).
Figure 6.
EMSA and supershift EMSA of the first RUNX site. Lane 1, wild-type Biotin-labeled probe alone; lane 2, wild-type probe + nuclear extract; lane 3, 100T/C probe + nuclear extract; lane 4, wild-type probe + nuclear extract + 300 fold unlabeled wild-type probe; lane 5, wild-type probe + rabbit anti-human RUNX3 antibody; lane 6, wild-type probe + nuclear extract; lane 7, wild-type probe + nuclear extract + anti-RUNX3 antibody; lane 8, wild-type probe + nuclear extract + normal rabbit IgG. Lower arrow indicates band shift upon formation of protein–DNA complex and upper arrow indicates the super-shifted complex in the presence of anti-RUNX3 antibody. One of four representative experiments is shown here.
RUNX3 regulates the ASP activity of L1Hs
An ASP activity that drives transcription of polII genes 5′ of L1Hs has been reported within +400 to +600 of the L1Hs 5′UTR (27,28). Since the third potential RUNX site fell within this region, we investigated whether it regulated transcription activated by the ASP. Wild-type or L1RP 5′UTRs mutated at the third RUNX site were inserted upstream of the luciferase gene in the antisense direction and the luciferase reporter assay was performed. Interestingly, the single-nucleotide mutations, 509T/C and 512T/A, which failed to have an effect in the sense direction, reduced ASP activity of the L1Hs 5′UTR (P = 0.061 and 0.002, respectively, Fig. 7A). In co-transfection experiments, ASP activity of the L1Hs 5′UTR was significantly decreased by 46% upon exogenous expression of dn-RUNX (P = 0.010, Fig. 7B), and increased by RUNX3 by ∼2 fold (P = 0.013). This latter effect of RUNX3 could be reduced by mutations at this site (P = 0.049 and 0.045, respectively). Again, RUNX1 and RUNX2 had no effect on the ASP activity.
Figure 7.
RUNX3 regulates the ASP activity of L1Hs 5′UTR. (A) Mutations, 509T/C and 512T/A, at the third RUNX site in the 5′UTR decrease the ASP activity of L1Hs. (B) Exogenous expression of RUNX3, but not RUNX1 or RUNX2, significantly increases the ASP activity, which is reduced by mutations at this site. dn-RUNX (AML1-ETO) significantly decreases the ASP activity (n = 9).
RUNX3 mRNA is expressed in many human tissues and certain cell lines
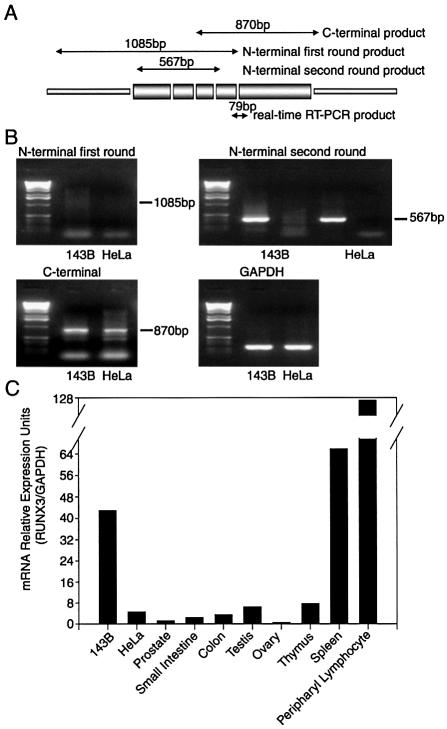
Since the above data indicated that RUNX3 was an important regulator of L1Hs transcription and retrotransposition, we studied its expression pattern in cell lines and human tissues. To examine whether full-length RUNX3 mRNA was indeed produced in 143B and HeLa cells, we performed RT–PCR as described previously (36) with primer pairs covering the N- and C-terminal regions of RUNX3, respectively (Fig. 8A). Due to the high-GC content in the N-terminal region of RUNX3, we used a nested PCR strategy to amplify the product efficiently. We found that full-length RUNX3 mRNA was present in both 143B and HeLa cells (Fig. 8B). To better quantify RUNX3 expression, we performed real-time quantitative RT–PCR using a pair of primers specific for human RUNX3, and found that RUNX3 was expressed at a much greater level in 143B cells than in HeLa cells (Fig. 8C), which may partially account for the higher L1Hs retrotransposition efficiency in 143B cells. RUNX3 was also expressed in all the human tissues tested (prostate, small intestine, colon, testis, ovary, thymus, spleen and peripheral lymphocyte), with the highest level in lymphoid tissues including peripheral lymphocyte, spleen and thymus. The RUNX3 level was also relatively high in testis.
Figure 8.
Expression of RUNX3 in cultured cells and human tissues. (A) Schematic figure of RUNX3 cDNA in which the relative positions of PCR primers and a 79 bp real-time RT–PCR product with primers (5′-ACTCAGCACCACAAGCCACT-3′ and 5′-GTCGGAGAATGGGTTCAGTTC-3′) are shown. (B) RT–PCR result of full-length RUNX3 mRNA expression in 143B and HeLa cells. A nested PCR strategy was performed to amplify the product at the N-terminal region of RUNX3 cDNA. Amplified product for the N-terminal region is only readily observed after second-round PCR, whereas amplified product for the C-terminal region is readily detectable. GAPDH is the internal control for sample loading. (C) RUNX3 mRNA is differentially expressed in 143B cells, HeLa cells and human tissues as revealed by real-time quantitative RT–PCR.
DISCUSSION
Here we report several lines of evidence indicating that the transcription factor, RUNX3, plays an important role in L1Hs transcription and retrotransposition. Mutations at the first RUNX site on the L1Hs 5′UTR significantly decreased protein interaction, transcription and retrotransposition. More importantly, exogenous expression of RUNX3 increased L1Hs transcription and retrotransposition, which were otherwise significantly decreased by RNAi targets against RUNX3 or dominant negative RUNX.
We used a luciferase reporter assay to demonstrate the regulatory role of RUNX3 in L1Hs transcription. In addition, we found that: (i) the promoter activity of L1RP 5′UTR was comparable with that of the SV40 promoter contained in pGL3-Control plasmid (Promega, data not shown), indicating that L1Hs 5′UTR harbors a very strong promoter activity; (ii) the promoter activity of L1RP 5′UTR was greater than that of L1.3 in both 143B and HeLa cells, which may partly explain the different retrotransposition activity of these two L1Hs elements; and (iii) transcription and retrotransposition of individual L1Hs were greater in 143B than in HeLa cells, which may partly result from the differential activation of the L1Hs 5′UTR in these two cell lines.
In the retrotransposition assay, exogenous expression of RUNX3 increased the L1Hs retrotransposition activity in 143B cells, but only by 15% (P = 0.104, Fig. 4B). This modest increase was probably due to a saturation effect of the endogenous RUNX3 that was highly expressed in this cell line (Fig. 8C). In fact, a significant effect of exogenous RUNX3 expression was observed in HeLa cells (P = 0.025, Fig. 4C), which express RUNX3 at much lower levels. It is also noteworthy that RUNX3 has only moderate transactivator activity on its own and stronger transactivation may require cooperation of other transcription factors (51). It is interesting to note that only one member of the RUNX family, RUNX3, played a regulatory role on L1Hs transcription and retrotransposition, and it is the most ancient of the three genes and represents the evolutionary founder of the mammalian RUNX family (52). However, exogenous expression of RUNX1 and RUNX2 also decreased L1Hs retrotransposition in 143B cells, although not in HeLa cells (Fig. 4B and C). Therefore, a suppressive role for RUNX1 and/or RUNX2 in a step after L1Hs transcription cannot be excluded. Indeed, study of the specific role of each RUNX factor in a cellular background lacking the other RUNX factors would be useful.
Using real-time quantitative RT–PCR, we found that 143B cells expressed a much higher level of RUNX3 than HeLa cells (Fig 8C), which may partially explain why 143B cells were particularly supportive of L1Hs retrotransposition. Although RUNX3 was expressed in all the tissues tested in our study, its expression has been reported to be mainly confined to hematopoietic cells (44); therefore, detection of RUNX3 in tissues such as colon and small intestine may be due to lymphocyte contamination. It is noteworthy that there was comparatively high RUNX3 expression in testis, and L1Hs retrotransposition has only been reported in transformed cells and germ cells (5). However, RUNX3 expression was unexpectedly low in adult ovary. Since L1Hs is actively transcribed in mouse ovary (53) and retrotransposition likely occurs in female germ cells (54), RUNX3 expression needs to be analyzed in fetal ovary. Recently, L1Hs has been successfully used to create a transgenic mouse model to study retrotransposition in vivo (53). Since RUNX factors are very well conserved in eukaryotic genomes (29), murine RUNX3 may facilitate retrotransposition of L1Hs in mouse germ cells. However, there are no obvious RUNX sites in the 5′UTR of the most active mouse L1 subfamily, TF (55) (data not shown). This suggests that the role of RUNX3 in L1 retrotransposition may be limited to certain species, such as humans.
Although the third RUNX site had no effect on L1Hs transcription in the sense direction, it resided in the region that possesses ASP activity (27). Therefore, we explored its potential regulation on the ASP activity of L1Hs 5′UTR. We found that the ASP activity of L1RP 5′UTR was 5–10-fold greater than background (cells transfected with promoterless pGL3-Basic vector, data not shown), which was consistent with Speek’s report (27); however, the L1RP ASP activity was only approximately 1/10–1/20 that of the sense promoter activity (data not shown). Binding of transcription factors to the ASP region has been proposed as a mechanism to regulate its activity (27). Here, we report for the first time a transcription factor, RUNX3, that can regulate the ASP activity of L1Hs 5′UTR, and possibly the subsequent expression of adjacent cellular genes in the human genome.
We employed a random mutagenesis strategy to identify important cis-elements for L1Hs retrotransposition. Most of our clones contained one to two mutations dispersed randomly in the mutagenized region, indicating that the method was efficient and successful. Among 50 randomly mutated clones tested to date, 31 had decreased retrotransposition activity by over 10% compared with L1RP (data not shown), suggesting that the 5′UTR indeed played an important role in L1Hs retrotransposition. Therefore, our study demonstrated the viability of a random mutagenesis strategy to study L1 retrotransposons, not only on the 5′UTR but also on other regions. In fact, random mutagenesis can provide a large number of L1 elements with readily identified mutations critical for L1 retrotransposition, which are extremely valuable to elucidate the mechanism underlying L1 retrotransposition. Furthermore, random mutagenesis mimics L1 evolution without natural selection, i.e. very active L1 elements that are usually lost due to a deleterious effect on the host are not eliminated in this in vitro system. Interestingly, we have found two clones out of 50 analyzed that contained single-nucleotide mutation and retrotransposed at ∼120% of the frequency of L1RP. Therefore, random mutagenesis may provide a powerful tool for directed evolution of L1, e.g. to create ‘hyperactive’ retrotransposons for practical applications.
In summary, our present study has demonstrated an important role for RUNX3 in L1Hs retrotransposition as well as transcription in both sense and antisense directions. Although many mobile elements have been widely used as genetic tools and even gene therapy delivery vehicles (56,57), the practical applications of L1Hs have been largely impeded by its relatively low and germ line-specific retrotransposition activity. Therefore, the present study should not only contribute to our understanding of the mechanism underlying L1Hs retrotransposition and its impact on the human genome, but may also, through the use of RUNX3 as a retrotransposition activator, facilitate the practical applications of L1Hs, e.g. in insertional mutagenesis and gene therapy.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Y. Groner for rabbit anti-human RUNX3 antibody and protocols for supershift EMSA, Y. Ito for eukaryotic expression plasmids of RUNX factors, and J. L. Goodier for pCEP4-puro plasmid. We thank T. R. Kadesch for helpful discussions and E. M. Ostertag and J. L. Goodier for critical reading of the manuscript. We also thank C. Sterner for assistance with luciferase assay and D. A. Ross for technical advice on EMSA. This work was supported by National Institutes of Health grant GM 045398.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Waterston R.H., Lindblad-Toh,K., Birney,E., Rogers,J., Abril,J.F., Agarwal,P., Agarwala,R., Ainscough,R., Alexandersson,M., An,P. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 3.Fanning T.G. and Singer,M.F. (1987) LINE-1: a mammalian transposable element. Biochim. Biophys. Acta, 910, 203–212. [DOI] [PubMed] [Google Scholar]
- 4.Brouha B., Schustak,J., Badge,R.M., Lutz-Prigge,S., Farley,A.H., Moran,J.V. and Kazazian,H.H.,Jr (2003) Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA, 100, 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostertag E.M. and Kazazian,H.H.,Jr (2001) Biology of mammalian L1 retrotransposons. Annu. Rev. Genet., 35, 501–538. [DOI] [PubMed] [Google Scholar]
- 6.Moran J.V. and Gilbert,N. (2002) Mammalian LINE-1 retrotransposons and related elements. In Craig,N., Craggie,R., Gellert,M. and Lambowitz,A. (eds), Mobile DNA II. ASM press; Washington, DC. pp. 836–869. [Google Scholar]
- 7.Hohjoh H. and Singer,M.F. (1996) Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J., 15, 630–639. [PMC free article] [PubMed] [Google Scholar]
- 8.Martin S.L. (1991) Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol., 11, 4804–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hohjoh H. and Singer,M.F. (1997) Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J., 16, 6034–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Q., Moran,J.V., Kazazian,H.H.,Jr and Boeke,J.D. (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell, 87, 905–916. [DOI] [PubMed] [Google Scholar]
- 11.Mathias S.L., Scott,A.F., Kazazian,H.H.,Jr, Boeke,J.D. and Gabriel,A. (1991) Reverse transcriptase encoded by a human transposable element. Science, 254, 1808–1810. [DOI] [PubMed] [Google Scholar]
- 12.Moran J.V., Holmes,S.E., Naas,T.P., DeBerardinis,R.J., Boeke,J.D. and Kazazian,H.H.,Jr (1996) High frequency retrotransposition in cultured mammalian cells. Cell, 87, 917–927. [DOI] [PubMed] [Google Scholar]
- 13.Luan D.D., Korman,M.H., Jakubczak,J.L. and Eickbush,T.H. (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell, 72, 595–605. [DOI] [PubMed] [Google Scholar]
- 14.Kazazian H.H. Jr and Goodier,J.L. (2002) LINE drive. retrotransposition and genome instability. Cell, 110, 277–280. [DOI] [PubMed] [Google Scholar]
- 15.Moran J.V., DeBerardinis,R.J. and Kazazian,H.H.,Jr (1999) Exon shuffling by L1 retrotransposition. Science, 283, 1530–1534. [DOI] [PubMed] [Google Scholar]
- 16.Boeke J.D. (1997) LINEs and Alus—the polyA connection. Nature Genet., 16, 6–7. [DOI] [PubMed] [Google Scholar]
- 17.Esnault C., Maestre,J. and Heidmann,T. (2000) Human LINE retrotransposons generate processed pseudogenes. Nature Genet., 24, 363–367. [DOI] [PubMed] [Google Scholar]
- 18.Swergold G.D. (1990) Identification, characterization and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol., 10, 6718–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minakami R., Kurose,K., Etoh,K., Furuhata,Y., Hattori,M. and Sakaki,Y. (1992) Identification of an internal cis-element essential for the human L1 transcription and a nuclear factor(s) binding to the element. Nucleic Acids Res., 20, 3139–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skowronski J., Fanning,T.G. and Singer,M.F. (1988) Unit-length line-1 transcripts in human teratocarcinoma cells. Mol. Cell. Biol., 8, 1385–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathias S.L. and Scott,A.F. (1993) Promoter binding proteins of an active human L1 retrotransposon. Biochem. Biophys. Res. Commun., 191, 625–632. [DOI] [PubMed] [Google Scholar]
- 22.Becker K., Swergold,G., Ozato,K. and Thayer,R. (1993) Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum. Mol. Genet., 2, 1697–1702. [DOI] [PubMed] [Google Scholar]
- 23.Weis L. and Reinberg,D. (1997) Accurate positioning of RNA polymerase II on a natural TATA-less promoter is independent of TATA-binding-protein-associated factors and initiator-binding proteins. Mol. Cell. Biol., 17, 2973–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tchenio T., Casella,J.F. and Heidmann,T. (2000) Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res., 28, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales J.F., Snow,E.T. and Murnane,J.P. (2002) Environmental factors affecting transcription of the human L1 retrotransposon. I. Steroid hormone-like agents. Mutagenesis, 17, 193–200. [DOI] [PubMed] [Google Scholar]
- 26.Yu F., Zingler,N., Schumann,G. and Stratling,W.H. (2001) Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res., 29, 4493–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speek M. (2001) Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol., 21, 1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nigumann P., Redik,K., Matlik,K. and Speek,M. (2002) Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics, 79, 628–634. [DOI] [PubMed] [Google Scholar]
- 29.Ito Y. (1999) Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells, 4, 685–696. [DOI] [PubMed] [Google Scholar]
- 30.Levanon D., Negreanu,V., Bernstein,Y., Bar-Am,I., Avivi,L. and Groner,Y. (1994) AML1, AML2 and AML3, the human members of the runt domain gene-family: cDNA structure, expression and chromosomal localization. Genomics, 23, 425–432. [DOI] [PubMed] [Google Scholar]
- 31.Westendorf J.J. and Hiebert,S.W. (1999) Mammalian runt-domain proteins and their roles in hematopoiesis, osteogenesis and leukemia. J. Cell. Biochem., (Suppl. 32–33), 51–58. [DOI] [PubMed] [Google Scholar]
- 32.Okuda T., van Deursen,J., Hiebert,S.W., Grosveld,G. and Downing,J.R. (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell, 84, 321–330. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q., Stacy,T., Binder,M., Marin-Padilla,M., Sharpe,A.H. and Speck,N.A. (1996) Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl Acad. Sci. USA, 93, 3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okuda T., Nishimura,M., Nakao,M. and Fujita,Y. (2001) RUNX1/AML1: a central player in hematopoiesis. Int. J. Hematol., 74, 252–257. [DOI] [PubMed] [Google Scholar]
- 35.Ducy P., Zhang,R., Geoffroy,V., Ridall,A.L. and Karsenty,G. (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell, 89, 747–754. [DOI] [PubMed] [Google Scholar]
- 36.Li Q.L., Ito,K., Sakakura,C., Fukamachi,H., Inoue,K., Chi,X.Z., Lee,K.Y., Nomura,S., Lee,C.W., Han,S.B. et al. (2002) Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell, 109, 113–124. [DOI] [PubMed] [Google Scholar]
- 37.Levanon D., Bettoun,D., Harris-Cerruti,C., Woolf,E., Negreanu,V., Eilam,R., Bernstein,Y., Goldenberg,D., Xiao,C., Fliegauf,M. et al. (2002) The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J., 21, 3454–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inoue K., Ozaki,S., Shiga,T., Ito,K., Masuda,T., Okado,N., Iseda,T., Kawaguchi,S., Ogawa,M., Bae,S.C. et al. (2002) Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nature Neurosci., 5, 946–954. [DOI] [PubMed] [Google Scholar]
- 39.Inoue K., Ozaki,S., Ito,K., Iseda,T., Kawaguchi,S., Ogawa,M., Bae,S.C., Yamashita,N., Itohara,S., Kudo,N. et al. (2003) Runx3 is essential for the target-specific axon pathfinding of trkc-expressing dorsal root ganglion neurons. Blood Cells Mol. Dis., 30, 157–160. [DOI] [PubMed] [Google Scholar]
- 40.Taniuchi I., Osato,M., Egawa,T., Sunshine,M.J., Bae,S.C., Komori,T., Ito,Y. and Littman,D.R. (2002) Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell, 111, 621–633. [DOI] [PubMed] [Google Scholar]
- 41.Woolf E., Xiao,C., Fainaru,O., Lotem,J.D.R., Negrearu,V.Y.B., Brenner,O., Berke,G.D.L. et al. (2003) RUNX3 and RUNX1 are required for T cell development during thymopoiesis. Proc. Natl Acad. Sci. USA, 100, 7731–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y.W., Yasui,N., Ito,K., Huang,G., Fujii,M., Hanai,J., Nogami,H., Ochi,T., Miyazono,K. and Ito,Y. (2000) A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc. Natl Acad. Sci. USA, 97, 10549–10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostertag E.M., Prak,E.T., DeBerardinis,R.J., Moran,J.V. and Kazazian,H.H.,Jr (2000) Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res., 28, 1418–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levanon D., Brenner,O., Negreanu,V., Bettoun,D., Woolf,E., Eilam,R., Lotem,J., Gat,U., Otto,F., Speck,N. et al. (2001) Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech. Dev., 109, 413–417. [DOI] [PubMed] [Google Scholar]
- 45.Levanon D., Bernstein,Y., Negreanu,V., Ghozi,M.C., Bar-Am,I., Aloya,R., Goldenberg,D., Lotem,J. and Groner,Y. (1996) A large variety of alternatively spliced and differentially expressed mRNAs are encoded by the human acute myeloid leukemia gene AML1. DNA Cell Biol., 15, 175–185. [DOI] [PubMed] [Google Scholar]
- 46.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson JD., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwahn U., Lenzner,S., Dong,J., Feil,S., Hinzmann,B., van Duijnhoven,G., Kirschner,R., Hemberger,M., Bergen,A.A., Rosenberg,T. et al. (1998) Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nature Genet., 19, 327–332. [DOI] [PubMed] [Google Scholar]
- 49.Miyoshi H., Shimizu,K., Kozu,T., Maseki,N., Kaneko,Y. and Ohki,M. (1991) t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl Acad. Sci. USA, 88, 10431–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erickson P., Gao,J., Chang,K.S., Look,T., Whisenant,E., Raimondi,S., Lasher,R., Trujillo,J., Rowley,J. and Drabkin,H. (1992) Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood, 80, 1825–1831. [PubMed] [Google Scholar]
- 51.Shi M.J., Park,S.R., Kim,P.H. and Stavnezer,J. (2001) Roles of Ets proteins, NF-kappa B and nocodazole in regulating induction of transcription of mouse germline Ig alpha RNA by transforming growth factor-beta 1. Int. Immunol., 13, 733–746. [DOI] [PubMed] [Google Scholar]
- 52.Levanon D., Glusman,G., Bettoun,D., Ben-Asher,E., Negreanu,V., Bernstein,Y., Harris-Cerruti,C., Brenner,O., Eilam,R., Lotem,J. et al. (2003) Phylogenesis and regulated expression of the RUNT domain transcription factors RUNX1 and RUNX3. Blood Cells Mol. Dis., 30, 161–163. [DOI] [PubMed] [Google Scholar]
- 53.Ostertag E.M., DeBerardinis,R.J., Goodier,J.L., Zhang,Y., Yang,N., Gerton,G.L. and Kazazian,H.H. (2002) A mouse model of human L1 retrotransposition. Nature Genet., 32, 655–660. [DOI] [PubMed] [Google Scholar]
- 54.Brouha B., Meischl,C., Ostertag,E., de Boer,M., Zhang,Y., Neijens,H., Roos,D. and Kazazian,H.H.,Jr (2002) Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am. J. Hum. Genet., 71, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naas T.P., DeBerardinis,R.J., Moran,J.V., Ostertag,E.M., Kingsmore,S.F., Seldin,M.F., Hayashizaki,Y., Martin,S.L. and Kazazian,H.H. (1998) An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J., 17, 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yant S.R., Ehrhardt,A., Mikkelsen,J.G., Meuse,L., Pham,T. and Kay,M.A. (2002) Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat. Biotechnol., 20, 999–1005. [DOI] [PubMed] [Google Scholar]
- 57.Richardson P.D., Augustin,L.B., Kren,B.T. and Steer,C.J. (2002) Gene repair and transposon-mediated gene therapy. Stem Cells, 20, 105–118. [DOI] [PubMed] [Google Scholar]