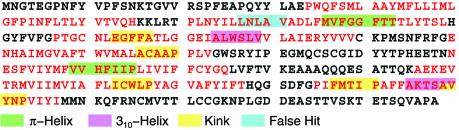
Figure 3.
Examining the impact of the parameter settings on the non-canonical conformations identified by the descriptor search engine in the bovine rhodopsin primary structure: helical regions are shown in red letters and non-helical regions in black. As shown in Table 3, reducing the system’s stringency increases the expected average amount of mislabeling. This is demonstrated here, again using the bovine rhodopsin sequence as the test input: for the least stringent combination of values in Table 3 (LogProbthres = –23 and P = 4), a kink is falsely predicted (shown in blue color), in addition to the correctly predicted non-canonical conformations. Notably, this is the only combination of values from Table 3 which will give rise to a false positive when processing the bovine rhodopsin sequence. See also text for a discussion.