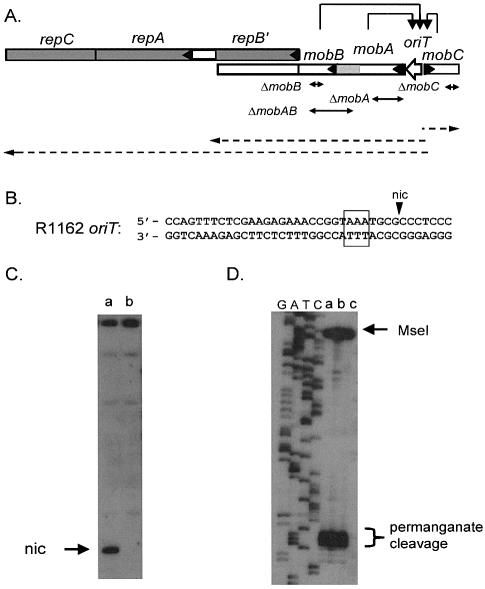
Figure 1.
(A) Region of R1162 containing the mob and rep genes (indicated by the open and filled rectangles, respectively, with an arrow head at the beginning of each gene). The product of repB′ is made as a fusion product with MobA, and is also translated separately (6). Thus, mobA is transcribed through mobB, but the two gene products are coded in separate reading frames. Transcripts are indicated by the dashed arrows. These transcripts originate from promoters located between oriT and mobC (6), and are negatively regulated by the proteins of the relaxosome (7), as indicated by the arrows. The approximate locations of the deletions in the mob genes are also shown. The patterned region of mobA encodes the region of the protein that interacts with MobB. (B) Base sequence of R1162 oriT, showing cleavage site (nic); bases within the box are sensitive to oxidation by permanganate. (C) Primer extension through oriT for (a) R1162, (b) pUT1579. The top strand in B was the template. (D) Primer extension on permanganate-treated template DNA, with the bottom strand of (B) as template. Plasmid DNAs were (a) R1162, (b) pUT1292, which contains a mutated oriT that forms relaxosome but is not cleaved (29 and S.Zhang, unpublished data) and (c) pUT1579. DNA was digested with MseI, which cleaves distal to the permanganate-sensitive region, prior to synthesis. On the left of the figure is a DNA dideoxy sequencing ladder generated from the oligonucleotide used in the primer extension.