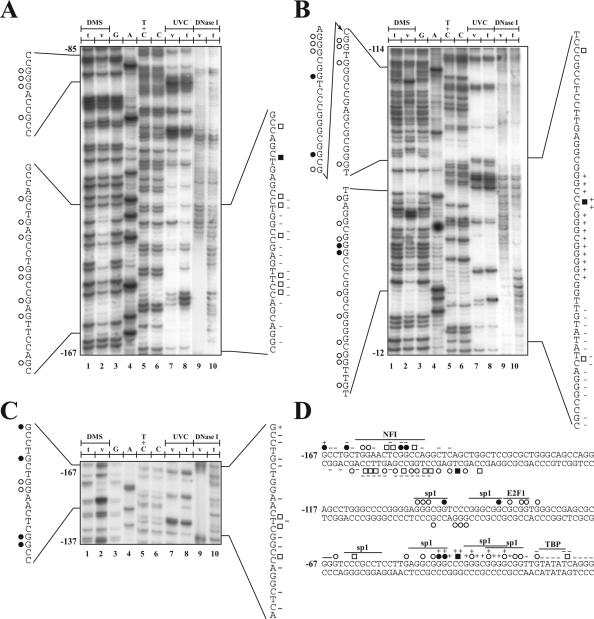
Figure 1.
Genomic footprinting of the human p21 gene promoter. The region shown was analyzed with primer set 1, to reveal the bottom strand sequence from nt −167 to −85 (A), and the primer set 2, to reveal the upper strand sequence from nt −114 to −12 (B) and nt −167 to −137 (C) relative to the transcription initiation site. Lane 1, 8 and 10: LMPCR of naked DNA purified from primary cultures of HSFs treated in vitro (t) with DMS (lane 1), UVC (lane 8) or DNase I (lane 10). Lanes 2, 7 and 9: LMPCR of DNA purified from HSFs treated in vivo (v) with DMS (lane 2), UVC (lane 7) or DNase I (lane 9) prior to DNA purification. Lanes 3–6: Maxam-Gilbert sequencing. DMS protected and hypersensitive guanines are indicated by opened and closed circles, respectively, on each side of the autoradiograms, whereas UVC protected and hypersensitive sites are indicated by opened and closed squares. The DNase I protected and hypersensitive sites are indicated by − and +, respectively. (D) Summary of the in vivo DMS, UVC and DnaseI footprints identified along the −187 to −136 human p21 gene promoter. The position of the consensus sequence for the specified transcription factors is also indicated above the sequence.