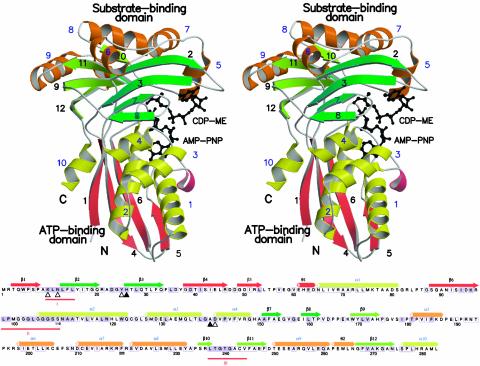
Fig. 3.
Sequence and structure of E. coli CDP-ME kinase. (Upper) Stereoview ribbon diagram of a monomer (β-strand numbers are black, and α-helix numbers blue). Strands and helices in the cofactor-binding domain are red and yellow, respectively. Strands associated with the two β-sheets in the substrate-binding domain are light and dark green, and the helices are orange. Substrate and AMP-PNP are depicted as black ball-and-stick models. N and C identify the amino and carboxyl termini. (Lower) Amino acid sequence of E. coli CDP-ME kinase with elements of secondary structure assigned and colored according to Upper. Residues strictly conserved or highly homologous in enzymes from S. typhi, Vibrio cholera, Haemophilus influenzae, Pseudomonas aeruginosa, Neisseria meningitidis, M. tuberculosis, Chlamydia trachomatis, and Clostridium perfringens are black on a purple background; when conserved in at least six of the sequences they are enclosed in a purple box. Sequences were retrieved from expasy (www.expasy.ch) and aligned (clustalw, ref. 63). ▵ identify residues interacting with the substrate using side chains; ▴ are for those using main chain. His-26 uses both. θ1 and θ2 identify 310-helices, and three red lines mark motifs I, II, and III.