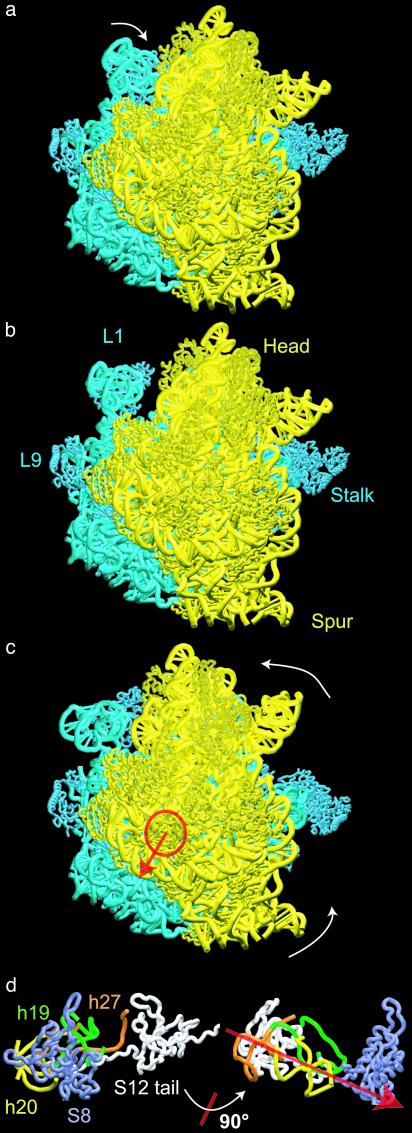
Fig. 1.
Structural rearrangements of the 70S ribosome obtained from elastic network NMA (30S subunit in yellow, 50S in blue). The ribosome is shown from the 30S solvent side. (a) The ribosome after a rearrangement along the first mode. The primary motion observed for this mode is displacement of the L1 protein to a position away from, or close to, the intersubunit space (also see Fig. 4b and Movies 1–3, which are published as supporting information on the PNAS web site). (b) The modified x-ray structure representing the equilibrium conformation of the ribosome. (c) The ribosome after displacement along mode 3. The primary motion is a rotation of the 30S subunit relative to 50S around the axis indicated in red. The amplitude of the displacement along this mode was adjusted to match the change due to the ratchet-like motion as observed in the cryo-EM maps of the structure during translocation; however, the direction of this displacement arose as a natural consequence of the NMA (see Supporting Text and Movies 1–3). (d)(Left) 30S RNA and protein components lying along the axis of rotation, viewed down that axis (close-up corresponding to the region indicated by the circle in c). (Right) Lineup of these components, viewed perpendicular to the axis (obtained from Left by a 90° rotation around an axis in the plane as indicated).