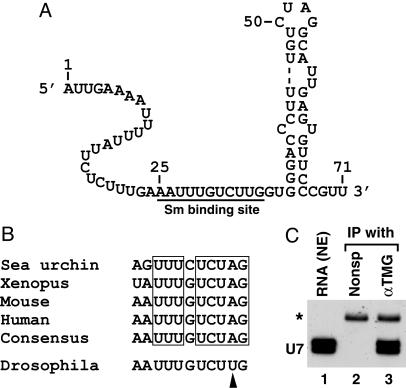
Fig. 4.
Features of Drosophila U7 snRNA. (A) The sequence and a possible secondary structure of 71-nt Drosophila U7 snRNA. The putative Sm-binding site is underlined. (B) Comparison of the Sm-binding site from four previously identified U7 snRNAs and from Drosophila U7 snRNA. The consensus sequence for the previously identified U7 snRNAs is shown and the invariant nucleotides in the U7 snRNA of all four species are boxed. The arrowhead indicates the nucleotide in Drosophila Sm-binding site that departs from the consensus. (C) RNA extracted from a Kc nuclear extract (lane 1) was incubated with either a nonspecific mAb (lane 2) or αTMG Ab (lane 3), and the RNA was recovered from protein G-Sepharose and analyzed by Northern blotting. A small amount of 32P-labeled 86-nt RNA (asterisk) was added to each sample before ethanol precipitation to control for the efficiency of RNA recovery.