Fig. 5.
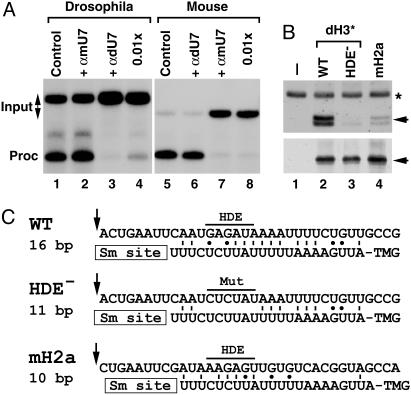
Drosophila U7 snRNA is present in histone pre-mRNA processing complexes. (A) In vitro processing of Drosophila dH3* (lanes 1–4) and mouse H2a-614 pre-mRNAs (lanes 5–8) in a Drosophila and mouse nuclear extract, respectively. Processing was carried out in the absence of any oligonucleotide (lanes 1 and 5) or in the presence of 2′-O-methyl oligonucleotides complementary to first 17 nt of Drosophila U7 snRNA (αdU7, lanes 3, 4, and 6) or mouse U7 snRNA (αmU7, lanes 2, 7, and 8). The oligonucleotides were used in the processing reaction at either 100 ng/μl (lanes 2, 3, 6, and 7) or 1 ng/μl (lanes 4 and 7). (B)A Drosophila nuclear extract was incubated in the absence of exogenous pre-mRNA (lane 1) or in the presence of three pre-mRNAs preannealed with the biotinylated adapter oligonucleotide: the WT Drosophila dH3*, its mutant containing 6-nt substitutions within the core HDE sequence (HDE–), and the mouse H2a-614 pre-mRNA. Bound Drosophila U7 snRNA and dSLBP, each indicated with the arrow, were analyzed by Northern blotting (Upper) and Western blotting (Lower). A small amount of 32P-labeled 86-nt RNA (asterisk) was added to each sample analyzed by Northern blotting. (C) A potential base pairing between the 5′ end of Drosophila U7 snRNA and the downstream element in three different pre-mRNAs analyzed in Fig. 5B. The arrow indicates a putative cleavage site located 4 nt after the stem (14) and the core purine-rich HDE beginning 13 nt after the cleavage site is overlined. The GU bp is indicated with the dot.