Abstract
Hormones and neurotransmitters rapidly change patterns of gene expression in target cells by activating protein kinases that phosphorylate and modify the activity of CREB and other transcription factors. Although CREB was initially characterized as mediating the response to cAMP, CREB phosphorylation and activation are stimulated by diverse extracellular signals and protein kinases in essentially all cells and tissues. CREB stimulates transcription through a constitutive activation domain (CAD), which interacts with the promoter recognition factor TFIID, and through a kinase-inducible domain (KID), when Ser-133 is phosphorylated. The present study provides new insight into the mechanism of activation by showing that each of the CREB domains contributes to transcription initiation by stimulating sequential steps in the transcription reaction. The CAD effectively assembled a polymerase complex, as evidenced by constitutive activation in vivo and stimulation of single-round transcription in vitro. In contrast, phosphorylation of the KID in CREB stimulated isomerization of the polymerase complex, as determined by abortive initiation, and promoter clearance and/or reinitiation, as measured by multiple rounds of transcription. Our results provide evidence for a new model for CREB-mediated induction through a concerted mechanism involving establishment of a polymerase complex by the CAD, followed by stimulation of isomerization, promoter clearance, and/or reinitiation by phosphorylated KID to enhance target gene transcription.
Important features of kinase-inducible gene regulation are that many of the target genes exhibit basal expression and the response to kinase activation is extremely rapid, occurring within minutes (1, 2). CREB was first described as a transcription factor mediating induction by extracellular signals activating adenylate cyclase and protein kinase A (PKA), which phosphorylates Ser-133 in CREB and enhances its transcriptional activity (3). Subsequently, several other signaling pathways were demonstrated to activate protein kinases that phosphorylate Ser-133 and enhance transcription of cAMP response element (CRE)-containing genes in a variety of cells and tissues (calmodulin kinase, mitogen-activated protein kinase/p90rsk, protein kinase C, p38, and protein kinase B (PKB/Akt) (4, 5).
For any gene to be transcribed, there must be recruitment of a polymerase complex, isomerization to expose the template strand, and promoter clearance of the polymerase to transcribe the body of the gene (6). Previous studies, based on the binding of heterogeneous activating transcription factor (ATF)/CREB proteins to CRE sites, have produced conflicting results (7–12). Early studies using footprinting demonstrated that the presence of an ATF/CRE site in a promoter produced an extended footprint, caused by a complex containing RNA polymerase II and TFIIB, TFIID, and TFIIE (7, 8). We previously showed that stimulation of in vitro transcription by cAMP-activatable PKA required a CRE site in the template and could be inhibited by a PKA-specific inhibitor peptide or by the addition of phosphatase but PKA did not affect binding of CREB (12). Additional in vitro transcription studies by others suggested that factors associated with ATF/CRE sites could promote any (7, 9, 10) or all (11) of the steps in transcription initiation in response to cAMP. However, interpretation of these studies is limited by the possible complication that PKA phosphorylates and changes the activity of other proteins in the nuclear extracts.
The CREB protein contains two distinct activation domains, a constitutive activation domain (CAD) and a kinase-inducible domain (KID) that can act independently to facilitate either constitutive or kinase-inducible transcription activation (13, 14). The CAD in CREB interacts with the hTAF130/dTAF110 subunit of TFIID, which may facilitate recruitment of a polymerase complex assembly (15, 16) (E. Felinski, J.K., L.J., and P.G.Q., unpublished work). We mapped the interaction between the TATA-binding protein (TBP)-associated factor (TAF) and CREB to hydrophobic residues in the CREB CAD (15). Subsequent work showed that these mutations also abolish (i) interaction between the proteins in vitro, (ii) recruitment of a polymerase-containing complex, and (iii) transcription activation in transfection studies, thereby providing a genetic link between interaction, recruitment, and transcription activation (E. Felinski and P.G.Q., unpublished work). An analogous mechanism is used to stimulate transcription activation by other constitutively active factors (17–20) and ligand-inducible nuclear receptors (21, 22).
Phosphorylation of CREB on Ser-133 has been demonstrated to promote association with CREB-binding protein (CBP). Bacterially expressed CREB binds CBP in a strictly phosphorylation-dependent manner and overexpression of CBP augmented induction of a somatostatin reporter by CREB in F9 cells (23). In that study, CBP also bound TFIIB, suggesting that it enhances activation through recruitment of essential components of the polymerase complex. In another study, CBP binding to CREB was reported in the absence of CREB phosphorylation (24). More recently, phosphorylated CREB was shown to interact with RNA polymerase complexes in HeLa cell nuclear extracts, an effect that could be blocked with a CBP peptide that bound CREB or antibody against RNA helicase A (25, 26). It was proposed that phosphorylated CREB activates transcription through interaction with CBP and recruitment of RNA polymerase II complexes. However, the assays used in that study did not assess the contribution of CREB phosphorylation to discrete steps in transcription initiation. Phosphorylated CREB would have to interact with the polymerase complex to have an effect on transcription, whether at the recruitment step or at later steps, e.g., isomerization, promoter clearance, or reinitiation.
In the present study, we used assays for discrete steps in transcription initiation to determine the separate and combined contributions of the CREB-activation domains, CAD and KID, to regulation of these different steps in transcription activation. Either or both of the CREB-activation domains were linked to the Gal4 DNA-binding domain in fusion proteins that were used to assess transcription activation in vivo and in vitro. We directly examined the contributions of these activation domains to recruitment, isomerization, or promoter clearance/reinitiation with single-round transcription, abortive initiation, or multiple-round in vitro transcription assays, respectively. We show that the CAD in CREB mediated assembly of a polymerase complex, a process that was unaffected by phosphorylation of KID. In contrast, the KID in CREB, but not the CAD, enhanced isomerization and multiple-round transcription in a phosphorylation-dependent manner. Although polymerase complex assembly by the CAD was not influenced by phosphorylation of KID, CAD-mediated assembly was required to observe enhancement of subsequent steps, indicating that the functions of these domains are integrated to provide maximal stimulation of the transcription initiation reaction in response to activation by protein kinases.
Materials and Methods
Transcription Assays.
JEG3 cells were transfected and luciferase activity, corrected for by the expression of the unregulated reporter, pRL-SV, was measured as before (27). For in vitro assays, recombinant CREB-Gal4 (CRG) proteins were expressed in baculovirus-infected Sf9 insect cells and purified by Gal4 DNA-binding site affinity chromatography. CRG proteins were quantitated by titration against a Gal4 probe of known specific activity in a mobility shift assay. Where indicated, 0.5 unit of PKA/0.5 μM okadaic acid were added to phosphorylate P-KID and P-CRG, after which 0.5 unit of PKA inhibitor was added to inhibit further PKA activity. Phosphorylation of CRG proteins by PKA was demonstrated by Western blotting with a CREB phospho-Ser-133-specific antibody. Rat liver nuclear extract preparation and in vitro transcription were performed as described (12), with the following modifications: in vitro reactions contained 50 μg of nuclear extract/150 fmol of template (750 fmol of Gal4 sites)/750 fmol of CREB-Gal4 protein. Template, nuclear extract, and CREB-Gal4 proteins were preincubated for 10 min to allow formation of a preinitiation complex before the addition of nucleotides. In vitro transcription reactions were performed for 10 min at 30°C. For single-round transcription, CTP and UTP were omitted for the initial 2 min and then added with 0.02% Sarkosyl, which prevents the formation of new preinitiation complexes and the reaction was continued for 10 min (28). Single- and multiple-round transcription depicted in Figs. 2 and 4 was assayed in parallel in three experiments, by preincubation of identical aliquots of template, nuclear extract, and CRG proteins, followed by the addition of the appropriate NTPs ± Sarkosyl. The mRNA synthesized was quantitated by primer extension analysis and was corrected for the relative amount of an internal RNA standard added in the transcription stop mix. For abortive initiation (29, 30), the nuclear extract was passed over Sephadex G50 to remove contaminating nucleotides (10). Abortive initiation reactions were performed in transcription buffer and included 1 mM dinucleotide substrate (ApG)/10 μM dATP/1 μM [α-32P]GTP, the next nucleotide to be added. The 32P-labeled mononucleotide substrate and trinucleotide product were separated on a 23% polyacrylamide/7 M urea gel. The results were normalized to the activity obtained with G4-DBD. Equivalent results were obtained for all in vitro assays by using at least two different preparations of purified proteins and nuclear extracts.
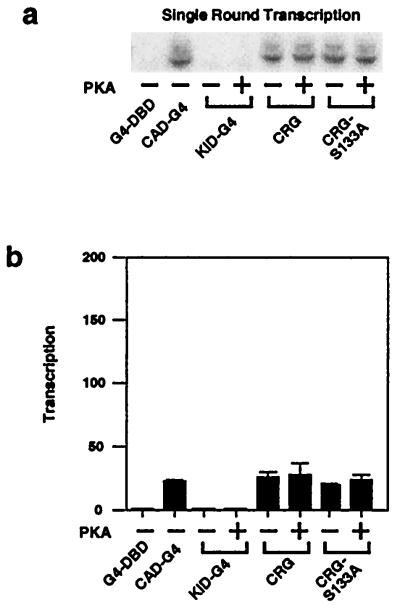
Figure 2.
The CAD, but not the KID, stimulates recruitment of a polymerase complex. Single-round transcription was measured by omission of two nucleotides until reinitiation was inhibited with 0.02% Sarkosyl after a 2-min preincubation. (a) A representative experiment is shown. (b) The results (means ± SEM) of four experiments are shown. These experiments and those in Fig. 4 were done in parallel with the same template, nuclear extract, and CREB-Gal4 proteins.
Figure 4.
Phosphorylation of CRG stimulates multiple-round transcription. (a) A representative in vitro transcription experiment for multiple-round transcription is shown (from the same experiment illustrating single-round transcription in Fig. 2a). (b) The results (means ± SEM) of four experiments are shown. (c) The number of cycles of transcription was estimated by dividing the multiple-round value (Fig. 4b) by the single-round value (Fig. 2b) for each protein. The results (means ± SEM) of four experiments are presented. These experiments and those in Fig. 2 were done in parallel with the same template, nuclear extract, and CREB-Gal4 proteins.
Results
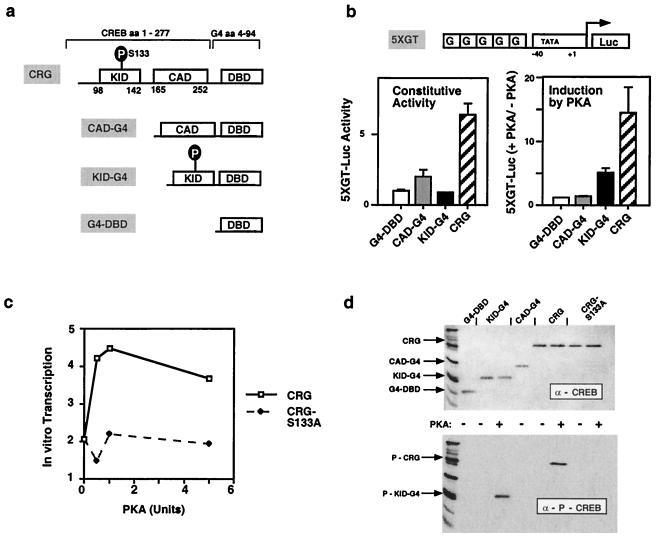
The activity of CREB-Gal4 fusion proteins, containing either (CAD-G4 or KID-G4) or both (CRG) of the CREB-activation domains (Fig. 1a), was assessed in transfected cells (Fig. 1b) with a minimal promoter under the control of five Gal4 sites (5XGT). The activity of 5XGT-Luc in the presence of G4-DBD is significantly greater than in mock-transfected cells, indicating that polymerase complexes do form at a low rate on the minimal promoter in the absence of activators. CAD-G4 or unphosphorylated CRG stimulated constitutive activity of the 5XGT promoter, but did not mediate induction by PKA (Fig. 1b). We and others have previously shown that unphosphorylated CRG and CRG-S133A are equally effective in promoting basal activity in transfected cells (13, 14). In contrast, cotransfection of the catalytic subunit of PKA with CRG or KID-G4 stimulated expression. The overall level of transcription induced by KID-G4 was reduced considerably and probably reflects stimulation of polymerase complexes that form in the absence of active recruitment. These results suggested that the two domains in CREB may affect different processes in transcription initiation, but analysis of this hypothesis required the establishment of an in vitro system in which the activities of these domains could be measured independently. An in vitro transcription system that recapitulates the regulation seen in vivo was developed by employing a Gal4-dependent promoter (5XGT), recombinant CREB-Gal4 fusion proteins (CAD-G4, KID-G4, and CRG) purified from Sf9 insect cells, and rat liver nuclear extracts containing RNA polymerase II and general transcription factors. As in vivo, induction of in vitro transcription was dependent on phosphorylation of CREB Ser-133 (CRG vs. CRG-S133A; Fig. 1c). Phosphorylation of the purified recombinant proteins was demonstrated by incubation with PKA and analysis of aliquots on parallel blots probed with antibody for CREB (Fig. 1d Upper) or phospho-Ser-133-CREB (Fig. 1d Lower). In vitro phosphorylation by PKA was specific for Ser-133 in CREB. Similar blots showed that phosphorylation of CRG/KID was maintained throughout the course of these assays (data not shown).
Figure 1.
Distinct activities of the CAD and KID in CREB in phosphorylation-dependent transcription activation in vivo and in vitro. (a) CAD, KID, and DNA-binding domain each contain amino acids 1–8 of CREB plus the indicated CREB amino acid sequence fused to amino acids 4–94 of the Gal4 DNA-binding domain. (b) JEG3 cells were cotransfected with the 5XGT-Luciferase reporter plasmid plus expression plasmids for CREB-Gal4 proteins ± the catalytic subunit of PKA. The data represent the means ± SEM of five independent experiments. (c) A representative experiment shows the effects of PKA titration on in vitro transcription mediated by CRG or S133A. (d) Western blots of purified CREB-GAL4 proteins, incubated ∓ 1 unit of PKA, were performed with anti-CREB (Upper) or anti-P-CREB (Lower) antisera.
To determine whether the CAD and/or KID could stimulate assembly of a polymerase complex, we measured single-round transcription, which reflects primarily recruitment (Fig. 2). The template was preincubated with nuclear extract and recombinant proteins for 10 min to allow the formation of preinitiation complexes. Then, two of the four nucleotides required for transcription were added for 2 min to allow initiation of transcripts, after which 0.02% Sarkosyl was added to inhibit new complex formation, together with the remaining nucleotides to allow completion of transcripts initiated by recruited polymerase complexes (10, 28). CAD-G4, CRG, or CRG-S133A were equally effective in recruiting a polymerase complex to the promoter and stimulating transcript synthesis. On the other hand, KID-G4 did not promote detectable recruitment of a polymerase complex in this assay, regardless of its phosphorylation state. Thus, the CAD in CREB is sufficient to recruit a polymerase complex. Although the KID can interact with CBP and the polymerase complex in a phosphorylation-dependent manner, this interaction does not necessarily contribute to recruitment, as had been suggested (23, 25, 26, 31). Rather, these data suggest that phosphorylated CREB may stimulate a later step in the transcription initiation pathway.
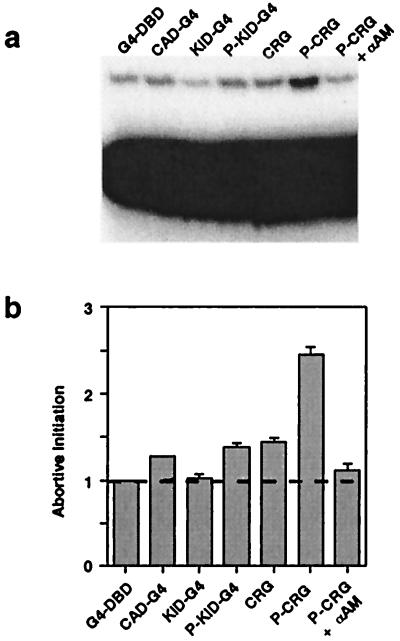
After recruitment, the RNA polymerase II complex undergoes an isomerization reaction, in which the promoter is melted to expose the template strand to the polymerase. The extent of this reaction can be measured by quantifying abortive initiation, i.e., the ability of the polymerase to catalyze addition of a nucleotide to a dinucleotide substrate complementary to the initiation site, which requires that the promoter be melted to expose the template strand (29, 30). The CAD-G4 and CRG proteins increased abortive initiation only modestly over the nonspecific background level provided by nuclear extract (Fig. 3 a and b), consistent with their ability to establish low-level basal transcription in vivo. In contrast, phosphorylation of CRG greatly enhanced the extent of abortive initiation, consistent with its ability to stimulate inducibility by PKA. Interestingly, phospho-KID-G4 alone stimulated abortive initiation poorly, which probably reflects its inability to recruit a polymerase complex (Fig. 2b). However, stimulation by CRG is greater because the CAD in CRG has recruited polymerase complexes, the isomerization of which can then be stimulated by phosphorylation of the KID in CRG.
Figure 3.
Phosphorylation of CRG stimulates abortive initiation. (a) The substrate and products of abortive initiation reactions were separated by electrophoresis. The nonspecific reaction was assessed by inclusion of 1 μg/ml α-amanitin to inhibit RNA polymerase II. (b) The combined results (means ± SEM) of four experiments are shown.
After recruitment and isomerization, the polymerase needs to break free of the preinitiation complex in a step referred to as promoter clearance, so that it can transcribe the body of the gene. Of the general transcription factors required to form the preinitiation complex, only TFIIF remains associated with the elongating polymerase (6, 32). On the other hand, it is likely that some factors, such as TFIID, TFIIB, and TFIIA, remain associated with the template and may promote reinitiation of the polymerase (33, 34). To determine the contributions of the CREB activation domains to promoter clearance and reinitiation, we measured multiple-round transcription, which reflects the summation of all steps in the transcription reaction (Fig. 4). The experiments shown in Figs. 2 and 4 were done in parallel with the same template, nuclear extract, and CREB-Gal4 proteins to allow direct comparison between the single- and multiple-round assays. CRG or CRG-S133A were more effective than CAD-G4 in stimulating multiple rounds of transcription, which is consistent with their greater effect in promoting constitutive activation in vivo (Fig. 1b). KID-G4 was ineffective in promoting multiple-round in vitro transcription, whether phosphorylated or not, probably as a result of its inability to recruit a complex in this assay (Fig. 2). In contrast, phosphorylated CRG stimulated multiple rounds of transcription (Fig. 4 a and b), consistent with phosphorylation affecting only the later steps in transcription. We also estimated the number of cycles of transcription (Fig. 4c) and found that the P-CRG-mediated increase in the number of cycles was greater than the extent of stimulation of isomerization by CRG (Fig. 3b). Thus, it seems likely that P-CRG stimulates promoter clearance and/or reinitiation, in addition to isomerization.
Discussion
The data presented here provide significant new insight into the mechanism of transcription activation used by phosphorylated CREB. Our results confirm and extend previous studies on the role of the CREB CAD in recruiting TFIID and a RNA polymerase complex to the targeted promoter. More importantly, our data provide new evidence for additional roles of phosphorylated CREB in promoting later steps in transcription activation. Thus, a revised model for the stimulation of transcription activation by phosphorylated CREB is proposed to involve recruitment of TFIID and the polymerase complex by the CAD, followed by stimulation of isomerization and promoter clearance and/or reinitiation by phosphorylation of the KID in CREB.
Previous work showed that the CAD in CREB can interact with TFIID (35), through association with hTAF130/dTAF110 (15, 16). A similar mechanism is involved in activation of transcription by other factors (17–22). In particular, disruption of the interaction between Sp1 and TAF130 disrupts transcription activation (16, 19). We recently showed that the CAD in CREB promotes recruitment of a complex containing RNA polymerase II, TFIIB, and TBP (E. Felinski, J.K., J.L., and P.G.Q., unpublished work) and that mutations in the CREB CAD that abolish the CREB-TAF interaction also abolish both recruitment and transcription activation (E. Felinski and P.G.Q., unpublished work). Here we show that the CAD in CRG is sufficient to recruit a polymerase complex and promote single-round transcription in vitro (Fig. 2). Taken together, these studies suggest that recruitment of TFIID and the polymerase complex is not only necessary, but sufficient, for the establishment of basal transcription by CREB. That idea is consistent with experiments showing that the association of holoenzyme with a promoter by a variety of means is sufficient to activate transcription (36, 37). In particular, our results are in excellent agreement with a recent report showing that recruitment of TFIID, but not holoenzyme, was sufficient to activate transcription in mammalian cells (38). Our results also provide evidence that the activity of the recruited complex can then be further stimulated by phosphorylation of CREB.
In previous work, Nakajima et al. (25, 31) assessed the ability of general transcription factors that bound to CREB immobilized on a column to support transcription activation when supplied with template and other factors. They found that TFIID was bound by immobilized CREB, independent of its phosphorylation state, and that a holoenzyme complex in nuclear extracts bound CREB in a phosphorylation- and CBP-dependent manner. Binding of both TFIID and the polymerase complex was required for maximal effects in the subsequent transcription assay. Those results were interpreted to indicate that the interactions of the CAD with hTAF130 and of phospho-CREB with CBP were responsible for recruiting TFIID and the holoenzyme complex, respectively. However, the experimental design used there would not allow observation of a processive mechanism of recruitment because template was provided only after protein–protein interaction between CREB and nuclear extract components (TFIID or holoenzyme fraction). In addition, phospho-CREB would have to interact with one or more components of the polymerase complex to affect any step in transcription initiation. Thus, the association of phospho-CREB with the polymerase complex does not, by itself, indicate that the phospho-CREB–CBP interaction regulates recruitment to the template. In our experiments, recruitment was measured in the presence of the template, and we saw that CAD, but not KID, mediated polymerase complex assembly. Although our data do not rule out a role for phospho-CREB-CBP in recruitment of a RNA polymerase complex, they do indicate that it is not essential for kinase-induced stimulation of transcription. A contribution of phospho-KID to isomerization, promoter clearance, and reinitiation would not have been evident in the experiments of Nakajima et al. (25). A consistent feature of studies from both laboratories is the necessity for recruitment of a TFIID complex by the CAD.
A significant new finding of this study is that our data clearly show that CREB facilitated postrecruitment steps in the transcription initiation reaction in a strictly phosphorylation-dependent manner. Thus, even if phospho-CREB did contribute to recruitment, its stimulation of later steps, which were less efficiently stimulated by the CAD, plays an essential role in promoting maximal transcription activation. Phosphorylation was required to see a large effect on isomerization or multiple-round transcription. There was no effect of phosphorylation on single-round transcription, indicating that later steps are not rate limiting under these conditions. In addition, the effect of phosphorylation on abortive initiation was less than the effect on multiple-round transcription, indicating that promoter clearance and/or reinitiation are also regulated. The results of our in vitro mechanistic studies (Figs. 2–4) correlate very well with the effects seen with the separate and combined domains of CREB in transfected cells (Fig. 1b). The ability of CAD-G4 to assemble a polymerase complex (Fig. 2) sets a basal level well below the maximal level seen with phosphorylated CRG, consistent with ineffective stimulation of later steps in the reaction (Figs. 3 and 4). In contrast, basal activity with KID-G4 alone was comparable to that seen with the inert G4-DBD, but allowed some induction by PKA in vivo, consistent with stimulation of later steps in the reaction for the few polymerase complexes that form on the template in the absence of activator-mediated recruitment.
We hypothesize that recruitment of TFIID by CAD is absolutely required to establish a polymerase complex, whose activity can then be modified by phospho-KID. A concerted mechanism of CREB-mediated transcription activation, involving regulation of sequential steps in the transcription reaction by the CREB constitutive and kinase-inducible domains, is consistent with many observations in different biological systems. The distinct roles of the CREB activation domains in transcription activation provides a basis for why CRE modulator (CREM) family members with large portions of the CAD spliced out act negatively as competitors (39) and why S133A acts as a dominant-negative factor (40, 41). Most importantly, it provides a mechanistic explanation for how neurotransmitters and hormones can rapidly increase the rate of transcription of target genes in response to extracellular signals (4). A well-documented example is cAMP induction of transcription of the phosphoenolpyruvate carboxykinase gene, which is dependent on CREB (27, 42, 43) and occurs within minutes of stimulation of cells with cAMP (1). In addition, mutation of the phosphoenolpyruvate carboxykinase CRE reduced both basal and kinase-inducible transcription of the gene (27, 42, 43). Regulatory regions of the phosphoenolpyruvate carboxykinase promoter corresponding to the CRE and TATA box were protected from DNase digestion independently of stimulation of the cells with cAMP, but only in cells expressing the phosphoenolpyruvate carboxykinase gene, not in cells where it is silent (26, 41–43). Constitutive occupancy of the CRE and promoter is consistent with the model proposed here, in which the effect of PKA phosphorylation of CREB is to change the activity of preexisting polymerase complexes, rather than promote their recruitment.
In summary, this work, together with previous work (13–15, 26, 31, 35), supports a general model for transcription activation in which CREB plays a role in regulating several steps in transcription initiation. CREB is constitutively bound to many promoters where, in concert with other factors, it will assemble a polymerase complex. This recruitment activity of the CAD is responsible for setting the basal activity of the gene and preparing it for responsiveness to external signals. Activation of protein kinases and phosphorylation of CREB in response to extracellular signals can then modify the activity of a preassembled polymerase complex by phosphorylating CREB to enhance isomerization, promoter clearance, and/or reinitiation of the polymerase, as shown here, leading to rapid increases in the transcription of target genes.
Acknowledgments
We thank E. Felinski, D. Spector, J. Hopper, and V. Chau for critical discussion of the work presented here. This work was supported by National Institutes of Health Grant R01 DK43871 (P.G.Q.).
Abbreviations
- CAD
constitutive activation domain
- KID
kinase-inducible domain
- CRG
CREB-Gal4
- PKA
cAMP-activatable protein kinase A
- CRE
cAMP response element
- CBP
CREB-binding protein
- 5XGT
minimal promoter controlled by five Gal4 sites
- G4-DBD
Gal4 DNA binding domain
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sasaki K, Cripe T P, Koch S R, Andreone T L, Petersen D D, Beale E G, Granner D K. J Biol Chem. 1984;259:15242–15251. [PubMed] [Google Scholar]
- 2.Montminy M R, Gonzalez G A, Yamamoto K K. Trends Neurosci. 1990;13:184–188. doi: 10.1016/0166-2236(90)90045-c. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez G A, Montminy M R. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 4.De Cesare D, Fimia G M, Sassone-Corsi P. Trends Biochem Sci. 1999;24:281–285. doi: 10.1016/s0968-0004(99)01414-0. [DOI] [PubMed] [Google Scholar]
- 5.Du K, Montminy M. J Biol Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 6.Zawel L, Reinberg D. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 7.Hai T W, Horikoshi M, Roeder R G, Green M R. Cell. 1988;54:1043–1051. doi: 10.1016/0092-8674(88)90119-5. [DOI] [PubMed] [Google Scholar]
- 8.Horikoshi M, Hai T, Lin Y S, Green M R, Roeder R G. Cell. 1988;54:1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 9.Narayan S, He F, Wilson S H. J Biol Chem. 1996;271:18508–18513. doi: 10.1074/jbc.271.31.18508. [DOI] [PubMed] [Google Scholar]
- 10.Wolner B S, Gralla J D. J Biol Chem. 1997;272:32301–32307. doi: 10.1074/jbc.272.51.32301. [DOI] [PubMed] [Google Scholar]
- 11.Narayan S, Beard W A, Wilson S H. Biochemistry. 1995;34:73–80. doi: 10.1021/bi00001a009. [DOI] [PubMed] [Google Scholar]
- 12.Brindle P, Linke S, Montminy M. Nature (London) 1993;364:821–824. doi: 10.1038/364821a0. [DOI] [PubMed] [Google Scholar]
- 13.Quinn P G. J Biol Chem. 1993;268:16999–17009. [PubMed] [Google Scholar]
- 14.Felinski E A, Quinn P G. J Biol Chem. 1999;274:11672–11678. doi: 10.1074/jbc.274.17.11672. [DOI] [PubMed] [Google Scholar]
- 15.Ferreri K, Gill G, Montminy M. Proc Natl Acad Sci USA. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill G, Pascal E, Tseng Z H, Tjian R. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coustry F, Sinha S, Maity S, deCrombrugghe B. Biochem J. 1998;331:291–297. doi: 10.1042/bj3310291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieberman P M, Berk A J. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 19.Saluja D, Vassallo M, Tanese N. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mengus G, May M, Carre L, Chambon P, Davidson I. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- 21.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 22.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 23.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. Nature (London) 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 24.Kee B L, Arias J, Montminy M R. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 26.Yeagley D, Agati J, Quinn P. J Biol Chem. 1998;273:18743–18750. doi: 10.1074/jbc.273.30.18743. [DOI] [PubMed] [Google Scholar]
- 27.Quinn P G, Granner D K. Mol Cell Biol. 1990;10:3357–3364. doi: 10.1128/mcb.10.7.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luse D S, Jacob G A. J Biol Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- 29.Goodrich J A, Tjian R. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 30.Lei L, Ren D, Finkelstein A, Burton Z F. Mol Cell Biol. 1998;18:2130–2142. doi: 10.1128/mcb.18.4.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 32.Zawel L, Kumar K P, Reinberg D. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 33.Yean D, Gralla J D. Nucleic Acids Res. 1999;27:831–838. doi: 10.1093/nar/27.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranish J A, Yudkovsky N, Hahn S. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing L, Gopal V K, Quinn P G. J Biol Chem. 1995;270:17488–17493. doi: 10.1074/jbc.270.29.17488. [DOI] [PubMed] [Google Scholar]
- 36.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 37.Gaudreau L, Adam M, Ptashne M. Mol Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- 38.Foulkes N S, Laoide B M, Schlotter F, Sassone C P. Proc Natl Acad Sci USA. 1991;88:5448–5452. doi: 10.1073/pnas.88.12.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Struthers R S, Vale W W, Arias C, Sawchenko P E, Montminy M R. Nature (London) 1991;350:622–624. doi: 10.1038/350622a0. [DOI] [PubMed] [Google Scholar]
- 40.Barton K, Muthusamy N, Fischer C, Clendenin C, Leiden J. Nature (London) 1996;379:81–85. doi: 10.1038/379081a0. [DOI] [PubMed] [Google Scholar]
- 41.Quinn P G, Wong T W, Magnuson M A, Shabb J B, Granner D K. Mol Cell Biol. 1988;8:3467–3475. doi: 10.1128/mcb.8.8.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xing L P, Quinn P G. Mol Endocrinol. 1993;7:1484–1494. doi: 10.1210/mend.7.11.8114762. [DOI] [PubMed] [Google Scholar]
- 43.Faber S, O'Brien R M, Imai E, Granner D K, Chalkley R. J Biol Chem. 1993;268:24976–24985. [PubMed] [Google Scholar]