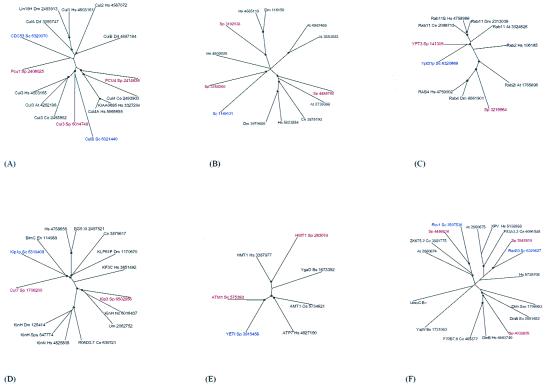
Figure 3.
Selected phylogenetic trees for protein families with apparent loss of paralogs in S. cerevisiae. (A) Cullins. (B) Cyclins (two cases of gene loss in S. cerevisiae). (C) Rab-type GTPase. (D) Kinesin ATPase subunit. (E) ABC transporter ATPase subunit. (F) DinB family of DNA repair polymerases. Circles denote nodes with at least 70% bootstrap support. The proteins are designated by their gene identifiers (GI numbers) in the nonredundant database and species abbreviations: Sc, S. cerevisiae; Sp, S. pombe; Nc, Neurospora crassa; Um, Ustilago maydis; Dm, D. melanogaster; Ce, C. elegans; Hs, Homo sapiens; Xl, Xenopus laevis; At, Arabidopsis thaliana; Os, Oryza sativa. Proteins from S. cerevisiae are color-coded blue, and those from S. pombe are coded red.