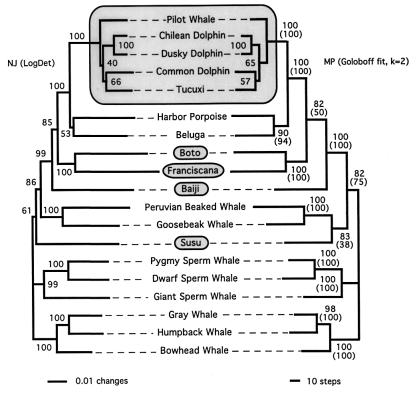
Figure 1.
Unrooted molecular phylogeny of cetaceans based on nuDNA and mtDNA sequences. The left and right sides show the NJ (under LogDet distances) and MP (under Goloboff fit criterion with k = 2) trees, respectively. Bootstrap values are indicated at the nodes (1,000 and 400 replicates for NJ and MP, respectively). Values between parentheses correspond to bootstrap values (400 replicates) for ML analyses (see Fig. 2 for tree topology). Topology of the MP tree is stable to Goloboff weighting (with k = 0–8). Unweighted MP analysis yielded a tree in which the positions of the sperm whale and [beaked whales + susu] clades are exchanged; these two alternative topologies have tree lengths differing by two evolutionary events and are not significantly different under the Kishino-Hasegawa test. The large and small shaded boxes indicate the radiation of delphinids and the four river dolphin species, respectively. Note that the exact placement of the susu differs between the NJ and MP/ML trees.