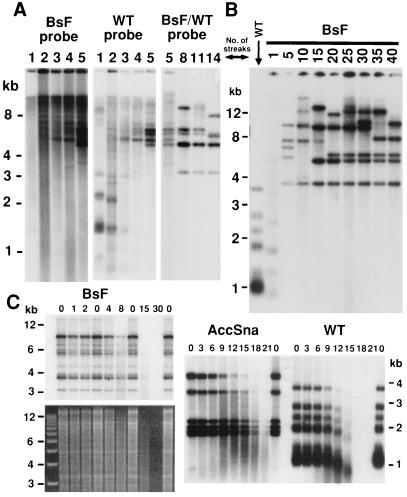
Figure 2.
Telomeric changes in ter1-BsF cells. (A) Southern blot of ter1-BsF mutant followed over 14 serial streaks after its creation (as numbered above lanes). EcoRI-digested genomic DNA was first hybridized with a probe specific to BsF mutant repeats (Left). After stripping, this filter was rehybridized with a probe specific to wild-type telomeric repeats (Center). This detects telomeres that have yet to have BsF repeats added onto them as well as telomeres elongated by the BsF mutant telomerase. Another filter with DNA from streaks 5, 8, 11, and 14 from the same clone probed with an oligonucleotide that binds equally to both wild-type and BsF repeats is shown (Right). The telomeric pattern of wild-type cells can be compared from B. (B) Southern blot showing telomeres of a single clonal lineage of ter1-BsF cells followed over 40 serial streaks on rich media. Genomic K. lactis DNA was digested with EcoRI and probed with a wild-type K. lactis telomeric oligonucleotide. DNA from the wild-type parental control is also shown. (C) Bal31 exonuclease time course done with DNA from ter1-BsF and ter1-AccSna mutants after formation of sharp bands carrying telomeric repeats. Numbers above lanes indicate length of Bal31 treatment in minutes. After exonuclease treatment, samples were digested with EcoRI before electrophoresis and Southern blotting. Hybridization was with a telomeric probe. Rate of disappearance of telomeric signal paralleled disappearance of total DNA in both mutants as seen on ethidium bromide straining of the gel, whereas disappearance of telomeric signal in wild type preceded complete digestion of total DNA (not shown).