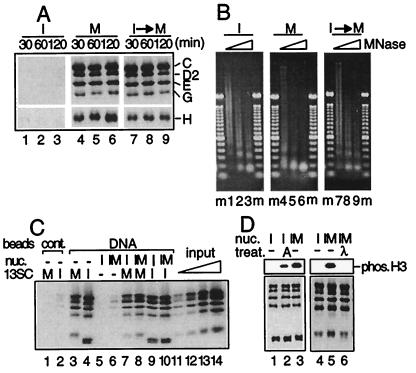
Figure 5.
Cell cycle regulation of interactions between 13SC and nucleosomes. (A) DNA-coupled magnetic beads were incubated with an interphase extract (lanes 1–3) or a mitotic extract (lanes 4–6) for different times as indicated. Alternatively, the DNA beads were first incubated with an interphase extract for 60 min and then with a mitotic extract for the indicated times (lanes 7–9). After washing the beads, bound proteins were analyzed by immunoblotting. (B) I nucleosomes, M nucleosomes, and I→M nucleosomes were assembled as shown in A. After washing the beads, DNA was digested with increasing amounts of micrococcal nuclease, isolated, and fractionated in a 1.25% agarose gel. m, 100-bp ladder marker. (C) DNA-coupled magnetic beads (lanes 4–10) or control beads (lanes 1 and 2) were incubated with buffer alone (lanes 1–4), with a condensin-depleted interphase extract for 120 min (lanes 5, 7, and 9), or with a condensin-depleted interphase extract for 60 min and then with a condensin-depleted mitotic extract for another 60 min (lanes 6, 8, and 10). The beads were washed and incubated with the mitotic form of 13SC (lanes 1, 3, 7, and 8), with the interphase form of 13SC (lanes 2, 4, 9, and 10), or with buffer alone (lanes 5 and 6) for 60 min. After washing the beads, bound 13SC was detected by immunoblotting. As standards, 6.25% (lane 11), 12.5% (lane 12), 25% (lane 13), and 50% (lane 14) of input 13SC were loaded in parallel. (D) Condensin-free I nucleosomes (lanes 1, 2, and 4) or I→M nucleosomes (lanes 3, 5, and 6) were assembled around DNA-coupled beads as described in C. The beads were washed and treated with protein kinase A (lane 2), λ phosphatase (lane 6), or control buffers (lanes 1 and 3–5). The phosphorylation states of histone H3 were analyzed by immunoblotting with a phosphospecific antibody that recognizes Ser-10 (Upper). Alternatively, the beads were incubated with the mitotic form of 13SC for 60 min. After washing the beads, bound proteins were analyzed by immunoblotting (Lower).