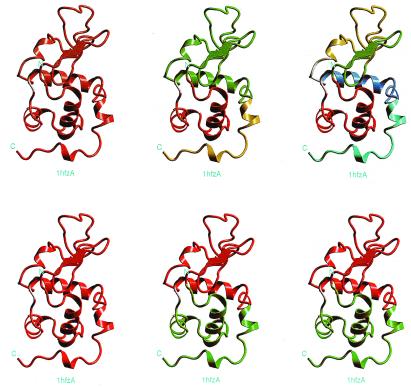
Figure 4.
The graphical representation at each anatomy level for α-LA (PDB code: 1hfzA). The results presented in this figure corresponds very nicely with the experimental results obtained by partial proteolysis (25). Note that, in this figure, a single helix constitutes a building block fragment. Because the scoring function is empirical, it is unclear above which threshold value a cut fragment is stable when isolated. According to the definition of the scoring function, if a fragment score is positive and the size is above 200 residues, the fragment will be more stable than at least half of the known conformations in the PDB for fragments with similar size (Fig. 2). A short helix assigned as a building block is not owing to its stability (score) but to its fragment size. If the size of an unassigned fragment is larger than the minimum size of a building block, it is assigned as a building block.