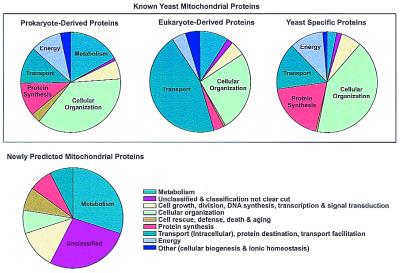
Figure 4.
Functions of yeast mitochondrial proteins are plotted for known mitochondrial proteins (upper three pie charts) and for the newly predicted mitochondrial proteins (lower pie chart). Each pie chart shows the percentage of proteins with a given function. Known mitochondrial proteins can be operationally divided into three populations: those with homologs in eubacteria or archaea (prokaryote-derived mitochondrial proteins), those with homologs only in other eukaryotes (eukaryote-derived mitochondrial proteins), and those without detectable homologs in the set of complete genomes (organism-specific mitochondrial proteins). Many functional systems, such as the mitochondrial ribosome, have components from more than one category of genes. The organism-specific mitochondrial proteins may be conserved in related species; many of the yeast-specific genes are conserved in other fungi as well, although absent in the more distantly related eukaryotes listed in Fig. 1A. Functional categories are defined as in the MIPS (Munich Information Center for Protein Sequences) database (29). For this analysis, mitochondrial proteins were predicted with an accuracy of 70% as scored by the self-consistency test.