Abstract
Peroxisomes are essential organelles of eukaryotic origin, ubiquitously distributed in cells and organisms, playing key roles in lipid and antioxidant metabolism. Loss or malfunction of peroxisomes causes more than 20 fatal inherited conditions. We have created a peroxisomal database (http://www.peroxisomeDB.org) that includes the complete peroxisomal proteome of Homo sapiens and Saccharomyces cerevisiae, by gathering, updating and integrating the available genetic and functional information on peroxisomal genes. PeroxisomeDB is structured in interrelated sections ‘Genes’, ‘Functions’, ‘Metabolic pathways’ and ‘Diseases’, that include hyperlinks to selected features of NCBI, ENSEMBL and UCSC databases. We have designed graphical depictions of the main peroxisomal metabolic routes and have included updated flow charts for diagnosis. Precomputed BLAST, PSI-BLAST, multiple sequence alignment (MUSCLE) and phylogenetic trees are provided to assist in direct multispecies comparison to study evolutionary conserved functions and pathways. Highlights of the PeroxisomeDB include new tools developed for facilitating (i) identification of novel peroxisomal proteins, by means of identifying proteins carrying peroxisome targeting signal (PTS) motifs, (ii) detection of peroxisomes in silico, particularly useful for screening the deluge of newly sequenced genomes. PeroxisomeDB should contribute to the systematic characterization of the peroxisomal proteome and facilitate system biology approaches on the organelle.
INTRODUCTION
Peroxisomes were first identified by de Duve in 1966 (1). Only a few months ago, the open debate on its ontogenetic as well as evolutionary origin has been settled: the organelle is derived from ER-membranes (2–4). This constitutes perhaps, a modern illustration in an organelle context, of the ‘ontogeny recapitulates phylogeny’ principle. Peroxisomes are indispensable for development, morphogenesis and differentiation, and play roles of paramount importance in hydrogen peroxide detoxification and fatty acid metabolism, hallmark functions of the organelle. The plasticity of their protein composition and biochemical function is remarkable and may vary according to the organism, cell type and/or environmental condition. Depending on species, peroxisomes are playing key roles on the degradation of amino acids, methanol and purines, or in the synthesis of bile acids, essential polyunsaturated fatty acids or penicillin [see reviews (5–7)]. Peroxisomes belong to the microbody family jointly with glyoxysomes of plants and glycosomes of trypanosomes, which are related peroxisomes specialized in glycolisis and glyoxylate pathways, respectively. In higher eukaryotes, peroxisomes play a key role in lipid homeostasis, adapting its copy number to obey cellular requirements, such as cold exposure, nutrients or drugs. At a first glimpse, some peroxisomal routes such as the β-oxidation of fatty acids could seem to be redundant with the mitochondria β-oxidation, but they are in fact complementary as there is substrate specificity between the two organelles. The peroxisomal β-oxidation is uncoupled of ATP synthesis thus leading to heat production in thermogenesis.
Gaining knowledge on peroxisome components, metabolic functions in the different organisms and their key players remains an important challenge. We wish to contribute to the field with PeroxisomeDB (http://www.peroxisomeDB.org). This relational database has been created joining expertise from peroxisome biologists and bioinformaticians, and provides a comprehensive and exhaustive reference list of peroxisomal proteins of human and Saccharomyces cerevisiae with extensive hyperlinks to databases, which we believe to be among the most useful currently available. In addition to combining these links on a single location, we have included useful tools for in silico detection of the organelle, based on the presence of four peroxins that we had recently found to be peroxisomal markers (2); and also for detection of peroxisome targeting signals (PTS) motifs for PTS1, PTS2 and for Pex19 binding sites, facilitating thereof the identification of novel candidate peroxisomal proteins.
RESULTS
The peroxisomal proteome
The available metabolic databases (i.e. Kegg) use to depict metabolic pathways regardless of the subcellular compartmentalization of the different enzymatic steps. PeroxisomeDB is helpful in this regard, because we integrate the different routes specifying which steps take place in peroxisomes. Further, current annotation of peroxisomal proteins in the databases is frequently incomplete and often does not distinguish between proteins, which have a confirmed peroxisomal sublocalization and those which are only candidates according to in silico predictions. Using annotated data derived from one of the most broadly used bio-ontology resources, Gene Ontology (GO), supplemented with curated experimental literature searches, we have built a complete peroxisomal proteome of Homo sapiens (encoded by 85 genes) and S.cerevisiae (encoded by 61 genes). To this core ensemble, we have added the peroxisomal proteins from the model organisms Mus musculus or Yarrowia lipolytica which lack their human orthologues, for instance the mouse urate oxidase or the several Pex that are mostly restricted to Y.lipolytica (Table 1).
Table 1.
PeroxisomeDB contents
| Database contents | Number of entries |
|---|---|
| Peroxisomal genes | 157 |
| Homo sapiens [GO:77(6)] | 85 |
| Saccharomyces cerevisiae (GO:51) | 61 |
| Present in M.musculus and not in human | 7 |
| Present in Y.lipolytica and not in human | 4 |
| Functions and metabolic pathways | 50 |
| Diseases | 22 |
| Interactive metabolic pathway schemes | 6 |
| Peroxisomal tools | 2 |
| Peroxisome identification tool | |
| Target signal predictor |
Number of entries refers to the number of genes manually annotated in peroxisomeDB. Gene Ontology (GO) followed by number indicates the number of peroxisomal entries previously annotated as such in GO; the number in brackets indicates misannotated entries identified in GO and not included in peroxisomeDB.
From gene to pathway, from metabolite to disease
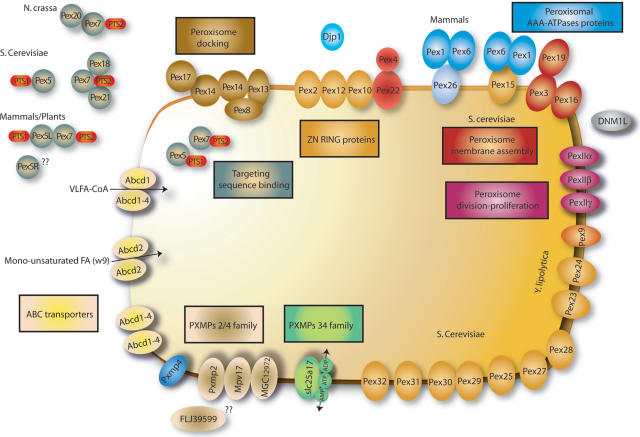
The proteomes are organized in interrelated sections: ‘Genes’, ‘Functions’, ‘Diseases’ and ‘Metabolic pathways’. The information is integrated in a section called ‘The Peroxisome at a Glance’, which includes four interactive schemes depicting the main peroxisomal functions: Lipid Metabolism, Glyoxylate and Dicarboxylate Metabolism, Amino acid and Purine Degradation and Antioxidant Metabolism. Two additional schemes are focused on the Peroxins and other peroxisomal membrane proteins (PMP) (Figure 1), and on Peroxisomal Disease.
Figure 1.
One of the six interactive schemes displayed in PeroxisomeDB: ‘Peroxins and other PMPs (peroxisome membrane proteins)’.
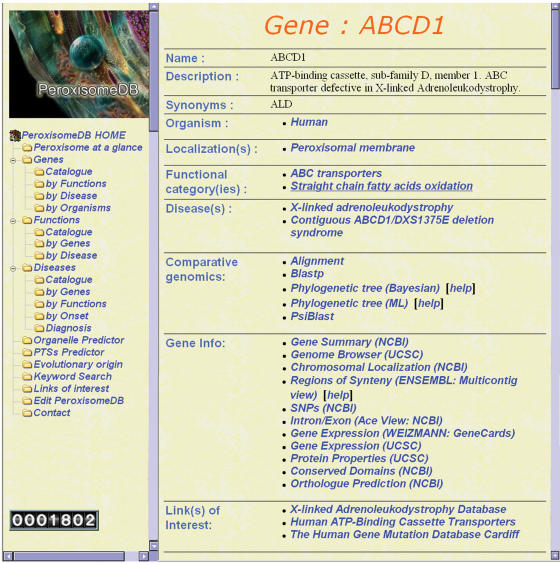
In the ‘Genes’ section, we have included: (i) a description and localization of the individual protein, (ii) its functional roles, (iii) the corresponding disease caused by protein malfunction, (iv) selected links to reference databases (Gene Info). Of particular relevance, we found the following sections: (i) from NCBI, the gene summary, the chromosomal localization, the predicted intron/exon structure at ACE VIEW, the Single Nucleotide Polymorphism (SNP) collection, the ortholog prediction and the conserved domains section; (ii) from ENSEMBL, the gene summary page that includes information on regions of syntheny; (iii) from the UCSC, the Proteome and Genome Browser with extensive expression data derived from microarray experiments; (iv) from the Weizmann Institute, the gene summary page with focus on expression data derived from microarrays, electronic northern and SAGE (Figure 2). Based on the biochemical and physiological context of protein function, we have organized the retrieved information in 50 different Functional Categories (Table 2). Noteworthy, 54 proteins belong to the lipid metabolism and 10 to the antioxidant categories.
Figure 2.
Gene Page for a given peroxisomal gene, including a brief description, localization, functional role, disease caused by malfunction if any, tools for functional genomics and selected links to reference databases (Gene Info).
Table 2.
Classification of the peroxisomal proteome into 50 different functional categories; the number of proteins classified according to functional category is listed
| Functional categories | Number of entries |
|---|---|
| Metabolism | 87 |
| Antioxidant | 10 |
| Antiinflammatory-antimicrobial | 1 |
| Catalases | 2 |
| Epoxides/Isochorismatase hydrolases | 1 |
| Gluthatione peroxidase/Thioredoxins | 1 |
| Microsomal detoxification system related | 1 |
| Peroxiredoxins | 2 |
| Superoxide dismutases | 2 |
| Glycerol synthesis | 1 |
| Glyoxylate and dicarboxylate metabolism | 10 |
| Lipid metabolism | 54 |
| Etherlipid and plasmalogen synthesis | 4 |
| Fatty acid oxidation | 24 |
| Branched chain fatty acid beta-oxidation | 9 |
| alpha-oxidation | 4 |
| Branched chain fatty beta-oxidation | 4 |
| di-trihydroxycholestanoic acid oxidation/bile acid | 6 |
| di-trihydroxycholestanoic acid beta-oxidation | 3 |
| Long-chain dicarboxylic acid oxidation | 5 |
| Straight chain fatty acid oxidation | 13 |
| Straight chain fatty acid beta-oxidation | 7 |
| Fatty acid synthesis/PUFA synthesis | 14 |
| Fatty acid chain elongation | 1 |
| Unsaturated fatty acid beta-oxidation | 12 |
| Long/very fatty acid activation | 9 |
| Regulation of acyl-CoA /CoA ratio | 11 |
| Nicotinate and nicotinamide metabolism | 3 |
| Protein/Amino acid metabolism | 10 |
| d-amino acid degradation | 2 |
| l-Lysine degradation | 3 |
| Polyamines degradation | 1 |
| Proteases | 2 |
| Transaminases | 2 |
| Purine metabolism | 2 |
| Retinoid metabolism | 1 |
| Peroxisomal membrane proteins (PMP) | 13 |
| ABC transporters | 6 |
| PXMP 2/4 family proteins | 5 |
| PXMP 34 family proteins | 2 |
| Peroxisome biogenesis proteins (peroxins) | 47 |
| Peroxisomal AAA-ATPases proteins | 4 |
| Peroxisomal division-proliferation | 12 |
| Peroxisome docking | 5 |
| Peroxisome matrix protein import | 13 |
| Zn Ring proteins | 6 |
| Peroxisome membrane assembly | 7 |
| Peroxisome targeting sequence binding | 8 |
| Peroxisome organization | 9 |
| Unknown | 3 |
The indispensable role of the organelle is stressed by the fatal consequences of the absence of peroxisomes in human diseases known as Peroxisome Biogenesis Disorders (PBD). Peroxisomal disorders are classified into two generic groups: (i) peroxisome biogenesis (PBD), (ii) single peroxisomal enzyme deficiencies (8). Mutations in peroxisomal proteins essential for biogenesis and matrix and membrane protein import (called peroxins or Pex genes) invariably lead to PBD: Zellweger Syndrome (ZS), Neonatal Adrenoleukodystrophy (NALD), Infantile Refsum disease (IRD) and Rhizomelic Chondrodysplasia Punctata (RCDP) are characterized by a strong reduction in number and size, or even complete absence of peroxisomes. Phenotypically, patients suffer from hepatic failure, neurodevelopmental delay, retinopathy and deafness with onset intrauteri or in the first months of life and fatal outcome within the first years (8). In the Disease Catalog, we have listed a total of 22 disorders with a brief disease description and links to Online Mendelian Inheritance in Man (OMIM) database, to specific disease-related mutation databases, to The Human Gene Mutation Database (HGMD) and to the SNPs database, that facilitate access to polymorphic markers useful for molecular genetic diagnosis. Of particular relevance for clinicians are new peroxisomal disorders, not described in previous reviews on peroxisomal disease. These include the contiguous ABCD1/DXS1375E deletion syndrome, and other disorders involving proteins which are not exclusively peroxisomal, such as malonyl-CoA decarboxylase (MLYCD) causing Malonic aciduria, xanthine oxidase (XDH) causing xanthinuria, acyl-CoA synthetase long-chain family member 4 (FACL4) for X-linked mental retardation, superoxide dismutase 1 (SOD1), mutated in amyotrophic lateral sclerosis (since 1% of SOD1 protein is located in peroxisomes) and fatty aldehyde dehydrogenase (FALDH) encoded by ALDH3A2, which has a splice variant of peroxisomal localization and causes Sjögren–Larsson syndrome.
Tools to identify peroxisomal proteins and peroxisomes
As an increasing amount of genomic sequences have become available, reverse genetics approaches have become very popular for identification of novel peroxisomal proteins, either by identifying consensus PTSs or by identification of orthologs of proteins determined to be peroxisomal in other species (homology probing). In PeroxisomeDB, we provide tools for facilitating both strategies, and also a simple and extremely useful approach for detecting the presence of the organelle in a given genome.
Comparative genomics tools
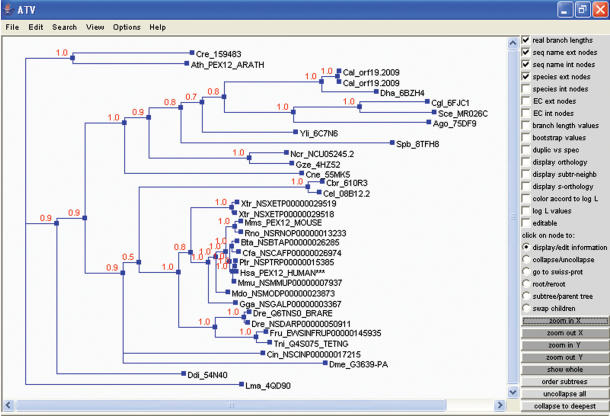
For facilitating identification of paralogs, orthologs and the main conserved domains throughout evolution, in-house developed routines provide the best BLAST and PSI-BLAST (9) results for each protein, in the form of precomputed homolog sequence results. Peroxisomal protein homologs with a region of similarity covering >50% of the query sequence are aligned using MUSCLE (10). Maximum Likelihood (ML) trees were built as implemented in PhyML (11). The tree built with the best-fitting model was further refined in a Bayesian analysis as implemented in MrBayes (12) (Figure 3). Both phylogenetic trees are provided in under PeroxisomeDB.
Figure 3.
Bayesian tree of a peroxisomal protein (PEX12) displayed in ATV [A Tree Viewer (21)]. Numbers at the nodes indicate the posterior probability of the corresponding partition. ***Indicate query sequence.
Peroxisomal targeting signal (PTS) predictor
Three different sequences targeting proteins to peroxisome have been identified and characterized to date (i) the 12 C-terminal residues PTS1 (13), (ii) N-terminal PTS2 for matrix proteins (14,15) and recently (iii) the Pex19 binding sites (Pex19 BS) for membrane proteins (16,17). We have manually selected the proteins carrying a PTS1 motif from 99 bona fide PTS1 proteins in H.sapiens, M.musculus, S.cerevisiae and Arabidopsis thaliana; also the PTS2 motif of the proteins ACAA1, PHYH and AGPS from H.sapiens, M.musculus, Ratus norvegicus, S.cerevisiae, Kluyveromyces lactis and A.thaliana; and for the Pex19 BS, we have chosen the best hits from yeast and humans (16,17). All these motifs have been experimentally characterized. On this manually curated selection of proteins carrying the PTSs, we have firstly applied Multiple EM for Motif Elicitation (MEME) (18) for refining a peroxisomal motif consensus. Then, we have submitted the three consensus PTS motifs to BLOCKS multiple alignment processor (19) in order to create three independent BLOCKS (sequence multiple alignment without gaps) of the highly conserved PTSs. Query sequence searches are carried out using ‘Do-It-YourSelf Block Search’ in BLOCKS server (http://blocks.fhcrc.org/blocks/). Results are returned with measures of probability, showing the pairwise alignment sequence (Figure 4). The resulting tool is a powerful PTSs Predictor, which integrates in a single location the three PTSs identification algorithms, and will allow for in silico identification of novel candidate peroxisomal proteins. This is of particular interest for PTS2 and PEX19BS containing proteins, since no web-based prediction tool is available for detection of those motifs.
Figure 4.
Target Signal Predictor. Tool for the prediction of the three peroxisomal target signals: PTS1, PTS2 and PEX19BS. Motifs within the query sequence are identified using ‘Do-It-YourSelf Block Search’ in BLOCKS server.
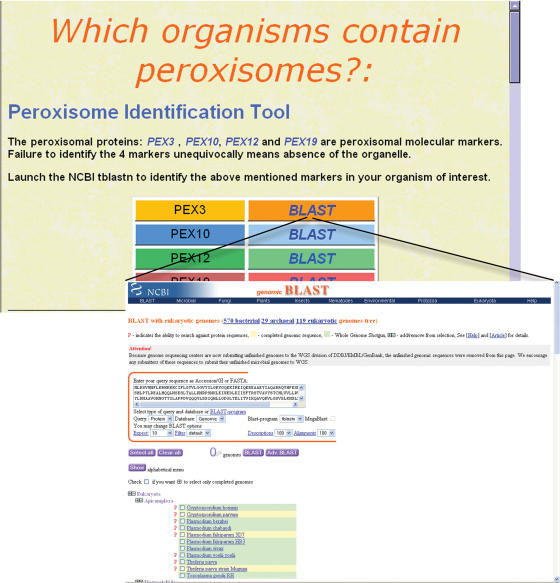
Peroxisome presence predictor
Detecting peroxisomes is not a trivial issue. Experimental peroxisome detection relies on direct visualization by electron microscopy or by immunohistochemistry against peroxisomal protein markers. Catalase is the most broadly used, although it can also be located in the cytosol. The common assumption that peroxisomes are present in all eukaryotic cells is contradicted, on experimental basis, in amitochondriates such as Encephalitozoon cuniculi, Giardia lamblia or Entamoeba histolytica (20). Using a molecular phylogenetic approach, we have very recently identified four membrane proteins as peroxisomal markers (Pex3, Pex10, Pex12 and Pex19) (2), what allowed us to demonstrate the absence of the organelle in the human pathogens Plasmodium falciparum or Toxoplasma gondii; parasites which do contain mitochondria and other organelles such as apicoplasts, micronemes or rhoptries. We have created a tool that allows automatic detection of peroxisomes by connecting the human sequences of the four marker peroxins to a page from NCBI that compiles an increasing amount of complete eukaryotic genomes (119 eukaryotic genomes to date in the site ‘Genomic Blast with eukaryotic genomes’, July 2006, Figure 5). Thus, direct blast searches to the most recent genomes and proteomes available, in the search for peroxisome presence becomes quick and simple. Absence of the organelle might provide insight into the metabolic status of an organism, and might even open new therapeutic strategies against human or animal pathogens. Indeed, using this tool we have substantiated our former findings by identifying another apicomplexa devoided of peroxisomes, the Theileria parva. Interestingly, the ciliates Tetrahymena thermophila and Paramecium tetraurelia do contain the markers. Ciliates and apicomplexa belong to the alveolata lineage, together with the dinoflagelata phylum, from which no complete genomes are yet available. Thus, it will be of interest to revisit the question of peroxisome loss within alveolata once new genomes become available.
Figure 5.
Peroxisome Identification Tool. It is based on the detection of four peroxisomal markers, by launching BLAST processes. The 119 different eukaryotic genomes provided by ‘genomic Blast with eukaryotic genomes page’ from NCBI, can be automatically blasted against the four markers.
Perspectives: maintenance and growth
Is our first priority to update PeroxisomeDB with new genomes and new peroxisomal entries curated manually by experts in biology of peroxisomes. Precomputed homology search results (BLAST and PSI-BLAST) will be automatically relaunched every 2 months. We have planned extensions to the available and annotated principal model organisms complete genomes such as mouse, rat and A.thaliana. In a second step, we will add the rest of vertebrate and fungi complete genomes, after inferring the components of their peroxisomal proteomes by homology probing and PTS prediction plus experimental literature when available. Most importantly, we wish to keep PeroxisomeDB highly dynamic by actively encouraging the users to submit their contributions through a web interface that allows automatic edition.
Acknowledgments
We gratefully acknowledge Jean Louis Mandel for scientific discussions, Sandra Metz for great graphic work, and the contribution of Elena Domínguez and Eva Barrio. This study was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the European Commission contract n° LSHM-CT2004-502987, the Association Française contre les Myopathies (Project n° 9315) the European Leukodystrophy Association and the Asociación Española contra la Leucodistrofia (ALE-ELA España). A.S. was a fellow of the AFM, Decrypthon Program and a recipient of FIS (CA05/0090) from the Instituto de Salud Carlos III. S.F. was a fellow of the European Leukodystrophy Association and the European Commission. Funding to pay the Open Access publication charges for this article was provided by Asociación Española contra la Leucodistrofia (ALE-ELA España).
Conflict of interest statement. None declared.
REFERENCES
- 1.De Duve C., Baudhuin P. Peroxisomes (microbodies and related particles) Physiol. Rev. 1966;46:323–357. doi: 10.1152/physrev.1966.46.2.323. [DOI] [PubMed] [Google Scholar]
- 2.Schluter A., Fourcade S., Ripp R., Mandel J.L., Poch O., Pujol A. The evolutionary origin of peroxisomes: an ER-peroxisome connection. Mol. Biol. Evol. 2006;23:838–845. doi: 10.1093/molbev/msj103. [DOI] [PubMed] [Google Scholar]
- 3.Gabaldon T., Snel B., van Zimmeren F., Hemrika W., Tabak H., Huynen M.A. Origin and evolution of the peroxisomal proteome. Biol. Direct. 2006;1:8. doi: 10.1186/1745-6150-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoepfner D., Schildknegt D., Braakman I., Philippsen P., Tabak H.F. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 5.Lazarow P.B. Peroxisome biogenesis: advances and conundrums. Curr. Opin. Cell Biol. 2003;15:489–497. doi: 10.1016/s0955-0674(03)00082-6. [DOI] [PubMed] [Google Scholar]
- 6.Titorenko V.I., Rachubinski R.A. The life cycle of the peroxisome. Nature Rev. Mol. Cell Biol. 2001;2:357–368. doi: 10.1038/35073063. [DOI] [PubMed] [Google Scholar]
- 7.Muller W.H., Bovenberg R.A., Groothuis M.H., Kattevilder F., Smaal E.B., Van der Voort L.H., Verkleij A.J. Involvement of microbodies in penicillin biosynthesis. Biochim. Biophys. Acta. 1992;1116:210–213. doi: 10.1016/0304-4165(92)90118-e. [DOI] [PubMed] [Google Scholar]
- 8.Wanders R.J. Metabolic and molecular basis of peroxisomal disorders: a review. Am. J. Med. Genet. 2004;126A:355–375. doi: 10.1002/ajmg.a.20661. [DOI] [PubMed] [Google Scholar]
- 9.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guindon S., Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 12.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 13.Neuberger G., Maurer-Stroh S., Eisenhaber B., Hartig A., Eisenhaber F. Motif refinement of the peroxisomal targeting signal 1 and evaluation of taxon-specific differences. J. Mol. Biol. 2003;328:567–579. doi: 10.1016/s0022-2836(03)00318-8. [DOI] [PubMed] [Google Scholar]
- 14.Glover J.R., Andrews D.W., Subramani S., Rachubinski R.A. Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo. J. Biol. Chem. 1994;269:7558–7563. [PubMed] [Google Scholar]
- 15.Mihalik S.J., Morrell J.C., Kim D., Sacksteder K.A., Watkins P.A., Gould S.J. Identification of PAHX, a Refsum disease gene. Nature Genet. 1997;17:185–189. doi: 10.1038/ng1097-185. [DOI] [PubMed] [Google Scholar]
- 16.Halbach A., Lorenzen S., Landgraf C., Volkmer-Engert R., Erdmann R., Rottensteiner H. Function of the PEX19-binding site of human adrenoleukodystrophy protein as targeting motif in man and yeast. PMP targeting is evolutionarily conserved. J. Biol. Chem. 2005;280:21176–21182. doi: 10.1074/jbc.M501750200. [DOI] [PubMed] [Google Scholar]
- 17.Rottensteiner H., Kramer A., Lorenzen S., Stein K., Landgraf C., Volkmer-Engert R., Erdmann R. Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol. Biol. Cell. 2004;15:3406–3417. doi: 10.1091/mbc.E04-03-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundy W.N., Bailey T.L., Elkan C.P. ParaMEME: a parallel implementation and a web interface for a DNA and protein motif discovery tool. Comput. Appl. Biosci. 1996;12:303–310. doi: 10.1093/bioinformatics/12.4.303. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiol. Rev. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zmasek C.M., Eddy S.R. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]