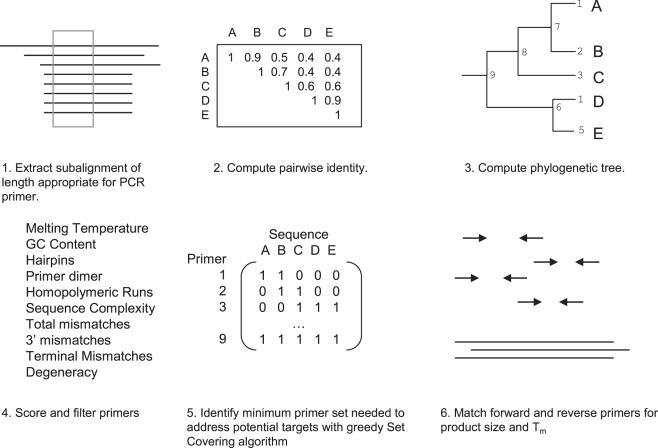
Figure 1.
Schematic diagram of SCPrimer design method. Sequences in a window the size of the desired primer (1) are compared to generate a similarity matrix (2), which is then used to build a phylogenetic tree (3). The consensus sequence for each branch of the tree is determined, then scored (4). Primers that do not pass the criteria are filtered out. A matrix corresponding to the ability of a primer to amplify a template is constructed, where 1 is true and 0 is false (5). The matrix is used by the set covering algorithm to determine the minimal set of primers required to amplify all sequences in the window. Primer pair candidates are matched for Tm and grouped by product size for user review.