Abstract
Methylating agents are widespread environmental carcinogens that generate a broad spectrum of DNA damage. Methylation at the guanine O6 position confers the greatest mutagenic and carcinogenic potential. DNA polymerases insert cytosine and thymine with similar efficiency opposite O6-methyl-guanine (O6MeG). We combined pre-steady-state kinetic analysis and a series of nine x-ray crystal structures to contrast the reaction pathways of accurate and mutagenic replication of O6MeG in a high-fidelity DNA polymerase from Bacillus stearothermophilus. Polymerases achieve substrate specificity by selecting for nucleotides with shape and hydrogen-bonding patterns that complement a canonical DNA template. Our structures reveal that both thymine and cytosine O6MeG base pairs evade proofreading by mimicking the essential molecular features of canonical substrates. The steric mimicry depends on stabilization of a rare cytosine tautomer in C·O6MeG–polymerase complexes. An unusual electrostatic interaction between O-methyl protons and a thymine carbonyl oxygen helps stabilize T·O6MeG pairs bound to DNA polymerase. Because DNA methylators constitute an important class of chemotherapeutic agents, the molecular mechanisms of replication of these DNA lesions are important for our understanding of both the genesis and treatment of cancer.
Keywords: crystal structure, DNA damage, DNA polymerase, protein–DNA complex
Alkylating agents are potent environmental carcinogens that are produced by burning tobacco or in grilling foods (1, 2) and also may be formed enzymatically in vivo (3, 4), for instance, by enzymatic metabolite nitrosation (5). Such agents cause a broad spectrum of DNA lesions. Although modifications at the O6 position of guanine constitute a minority of the total lesions, they are the most carcinogenic (6–8). The cytotoxic effects of DNA-methylating agents have been exploited in their use as potent anticancer agents. O6-methyl-guanine (O6MeG) is mutagenic because polymerases frequently misinsert T opposite O6MeG instead of C, both in vivo (9, 10) and in vitro (11–13). In this study, we present the crystal structures of complexes of a high-fidelity DNA polymerase with substrates representing several steps of nucleotide insertion opposite O6MeG. Additionally, we have engineered a substrate in which the O6MeG·C/T pair lies in DNA outside the binding site of the polymerase, allowing us to compare the conformation of these base pairs in duplex DNA to the conformation of the base pairs constrained in the polymerase active site.
The relative preference for incorporation of T and C opposite an O6MeG lesion varies somewhat with polymerase and sequence context (13). High-fidelity polymerases such as exonuclease-deficient E. coli polymerase I (Klenow fragment) (13) or bacteriophage T7 DNA polymerase (11) show an ≈7-fold preference for misinsertion of T. By comparison, these polymerases usually show a several thousand-fold preference for insertion of a correct base-pairing partner when copying a normal, undamaged DNA. O6MeG lesions slow the rate of DNA synthesis at the nucleotide incorporation step opposite the lesion (11–13) and subsequent extension beyond the O6MeG-pair (11, 12, 14). These kinetic effects vary with polymerase. Phage T7 polymerase shows 170- and 35-fold reductions in efficiency for nucleotide insertion and extension, respectively, as well as a preference for T misinsertion (11). In all polymerases studied to date, incorporation and extension for C and T·O6MeG base pairs is more efficient than for of any of the 12 possible unmodified base-base mismatches (11–13, 15).
High-fidelity DNA polymerases copy their templates rapidly and accurately. Structures of Bacillus stearothermophilus DNA polymerase I large fragment (BF) as it replicates undamaged (16–18) and damaged (19–21) DNA substrates yield a detailed picture of the structural mechanisms that ensure fidelity at each step of the reaction. An incoming template base first contacts the polymerase in an “open” state at a preinsertion site, where the template base is sequestered in a cleft between two α-helices in a high-syn conformation. This nucleotide subsequently is transferred to the insertion site and flipped into an anti conformation, with a concomitant enzyme conformational change into a “closed” state. This conformational change creates a tight binding site that is stereochemically complementary to the correct Watson–Crick base-pairing partner. This step has been reported to be critical for replication fidelity (22–25), but kinetic analyses of polymerases have shown that this may be an overly simplistic view of the reaction (26). After nucleotide triphosphate incorporation, the new base pair is translocated to a postinsertion site, and the system resets to an open conformation, with the next template nucleotide in the preinsertion site. Mispairs in the postinsertion site disrupt the polymerase active site; the extent of disruption correlates with observed reductions in mispair extension kinetics for other high-fidelity polymerases (16). Mismatch-mediated disruption of the polymerase active site provides a mechanism for ensuring fidelity past the initial incorporation step, because disrupted complexes tend to stall and disassociate rather than extend past the mispair. DNA mispairs up to four base pairs away from the primer terminus have been observed to disrupt the organization of the BF active site (16). DNA bound to polymerase undergoes significant conformational perturbations, including a transition from B form to A form DNA near the active site and the adoption of novel base-pairing schemes by several DNA mispairs (16). Changes in DNA hydrogen-bonding patterns indicative of the presence of rare tautomers, ionized bases, or other changes in the electronic character of these mispairs have not been directly observed.
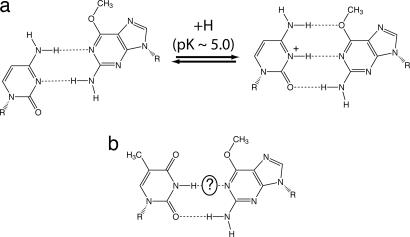
The structure and stability of DNA duplexes containing O6MeG·T or O6MeG·C base pairs have been characterized in the absence of polymerase. O6MeG·C forms a wobble pair in solution at neutral pH (27) (Fig. 1a). C·O6MeG pairs are more stable than the other three possible O6MeG pairings (but are weaker than canonical C·G pairings by 4–5 kcal/mol) (28). O6MeG·T mispairs are relatively unstable, forming only one hydrogen bond, but adopt a shape similar to Watson–Crick pairs (29, 30) (Fig. 1b). The Watson–Crick-like conformation of O6MeG·T mispairs has been postulated to allow preferential incorporation of T opposite O6MeG by high-fidelity polymerases (31) but cannot account for the fact that the polymerase only weakly discriminates against O6MeG·C. Here, we present nine high-resolution crystal structures that illustrate the extreme plasticity of O6MeG as a base-pairing partner. When bound to DNA polymerase O6MeG forms base pairs with both T and C that differ from the solution conformations of these base pairs and that mimic canonical, Watson–Crick base pairs.
Fig. 1.
Structures of O6MeG·C (a) and O6MeG·T (b) pairs in DNA duplexes, unbound by protein. Hydrogen bonds are shown as dashed lines. (a Left) O6MeG·C wobble pair from ref. 27. (a Right) Model of Watson–Crick O6MeG·protonated C pair from ref. 52. (b) O6MeG·T pairs from refs. 29 and 30. The presence or absence of the H bond indicated by the “?” has been the subject of some controversy (29, 30).
Results
We have determined pre-steady-state kinetic parameters for the insertion of C or misinsertion of T opposite a template G or O6MeG to contrast accurate and mutagenic replication of O6MeG by BF. Nine high-resolution crystal structures representing steps of O6MeG incorporation along the reaction pathway have been determined (Table 1 and supporting information (SI) Table 3): in the preinsertion site (+1 position), binding to an incoming ddCTP or ddTTP before chemistry in the closed conformation (0 position), after enzymatic incorporation in catalytically active BF crystals of a T or C opposite O6MeG (−1 base pair positions), and after extension of an additional correct base pair after T or C incorporation (−2 base pair position). For comparison, structures with O6MeG·T and O6MeG·C base pairs in duplex DNA in the solvent channels of our crystals (−10 base pair position) also have been determined.
Table 1.
Refinement resolution and R values
| Structure | OMG + 1 | OMGT 0 | OMGT − 1 | OMGT − 2 | OMGC 0 | OMGC − 1 | OMGC − 2 | OMGT − 10 | OMGC − 10 |
|---|---|---|---|---|---|---|---|---|---|
| PDB code | 2HHX | 2HHW | 2HW3 | 2HHV | 2HVH | 2HHU | 2HHT | 2HHQ | 2HHS |
| Resolution, Å | 50–2.26 | 50–1.88 | 50–1.98 | 50–1.55 | 50–2.49 | 50–1.80 | 50–2.05 | 50–1.80 | 50–1.80 |
| Completeness | 95.8 | 95.6 | 94.2 | 96.8 | 98.8 | 99.9 | 90.1 | 96.1 | 80.9 |
| Rwork / Rfree | 20.94/25.25 | 19.55/22.44 | 20.98/25.21 | 21.59/23.87 | 20.1/26.2 | 20.32/23.18 | 19.74/22.90 | 21.18/24.40 | 21.80/23.83 |
See SI Table 3 for additional statistics.
Kinetic Studies.
Using transient-state kinetic methods (32), we have determined pre-steady-state nucleotide-binding constants and polymerization rates for BF as it carries out accurate or mutagenic replication past a templating G or O6MeG (Table 2). As with other high-fidelity polymerases, BF shows a slight preference for misinserting T opposite O6MeG, ≈11-fold. Efficiency of insertion opposite O6MeG is ≈10-fold lower for BF than for T7 or Escherichia coli pol I enzymes (11, 13). The lower efficiency for O6MeG compared with G replication results from reductions in both the rate of nucleotide incorporation (kpol) and the binding constants for incoming nucleotide triphosphates (Kd).
Table 2.
Kinetic parameters for BF insertion opposite O6MeG
| Template | dNTP | KD, μM | kpol, s−1 |
kpol/KD, μM−1 s−1 |
Relative efficiency |
|---|---|---|---|---|---|
| G | dCTP | 17.9 | 193 | 10.8 | 1 |
| dTTP | 1.90 × 103 | 0.20 | 0.00011 | 9.8 × 10−6 | |
| O6MeG | dCTP | 1.10 × 103 | 0.12 | 0.00011 | 1.0 × 10−5 |
| dTTP | 368 | 0.46 | 0.00125 | 1.2 × 10−4 |
*Relative efficiency is the ratio of the kpol/KD to that for dCTP incorporation opposite G.
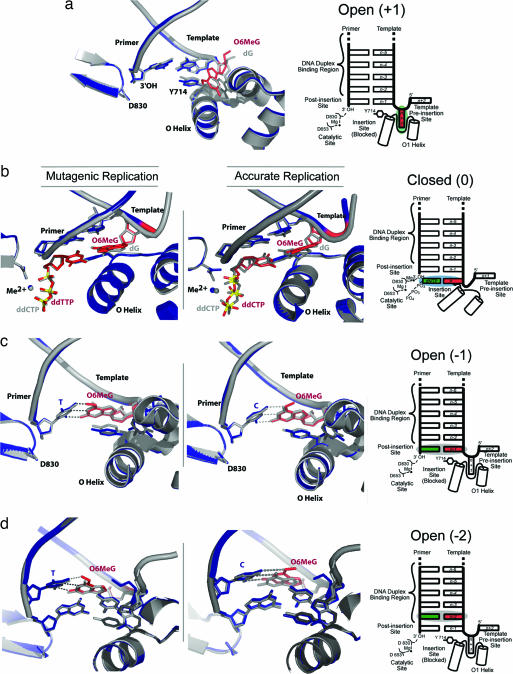
Preinsertion Site (+1 position).
We cocrystallized BF with a DNA substrate in which the next template base is an O6MeG (Fig. 2a). This structure resembles other preinsertion site complexes, with the O6MeG base resting positioned between the O1 and O helices. In unmodified DNA, the glycosidic bonds of nucleotides in this position are in the syn conformation, whereas O6MeG adopts an anti conformation to resolve the potential clash between the O6Me group and the walls of the preinsertion site. This change in conformation is restricted to the O6MeG base and sugar; the conformations of the protein and the duplex region of the DNA are unperturbed.
Fig. 2.
Structural snapshots of BF replicating past O6MeG lesions. (a Left) O6MeG in the template preinsertion site. The O6MeG-containing structure is shown in blue, with the O6MeG in red. The structure is superimposed on 1L3U, shown in gray. (b Left and Center) ddTTP·O6MeG (Left) and ddCTP·O6MeG (Center) pairs in the active site in the polymerase closed conformation superimposed on a ddCTP·G ternary complex structure and colored in the same manner as a. (c Left and Center) T·O6MeG (Left) and C·O6MeG (Center) pairs in the polymerase postinsertion site (−1 base pair position), superimposed on 1L5U. (d Left and Center) T·O6MeG (Left) and C·O6MeG (Center) pairs in the −2 base pair position, superimposed on 1L3S. (a–d Right) Schematic illustrations of the positions of the O6MeG lesions in a–d Left.
Insertion Site (0 Position).
We have captured a structure of BF in a closed conformation with a template O6MeG paired opposite an incoming ddTTP. This base pair is well ordered with good density for the methyl group. SI Fig. 6a shows the T·O6MeG pair from the ternary complex overlaid with composite-omit density for the base pair. The O6Me group points toward the Watson–Crick face and is interposed between the O6 and the thymine carbonyl oxygen (O4). The O6-methyl group in O6MeG·T or O6MeG·C base pairs has been observed in ref. 30 or modeled in refs. 27 and 29 with the methyl group pointing away from the Watson–Crick face. Comparison of the insertion site of the ddTTP·O6MeG complex (Fig. 2b Left, colored structure) with a normal, closed C·G structure (Fig. 2b Left, gray structure) shows that the damaged and undamaged base pairs adopt nearly identical conformations in the polymerase active site, except that the O6MeG is twisted slightly out of the plane of the thymine.
A ternary complex structure of BF with a templating O6MeG paired with an incoming ddCTP is similar in conformation to the T·O6MeG ternary complex and to a normal C·G pair (Fig. 2b Center). SI Fig. 6b shows the C·O6MeG pair with its composite omit density. Again, the O6 methyl group is interposed between the O6 of the methyl guanine base and the cytosine amine group.
Postinsertion Site (−1 Base Pair Position).
The consequences of replicating past O6MeG lesions can be observed directly because BF polymerase crystals retain catalytic activity. Soaking either dTTP or dCTP into BF:DNA cocrystals with O6MeG in the preinsertion site yields T·O6MeG or C·O6MeG pairs in the −1 base pair position (Fig. 2c Left and Center, respectively). At this site, these base pairs adopt a conformation that is essentially isosteric to BF complexes with undamaged bases. The C·O6MeG base pair adopts a conformation that is distinct from the wobble pair observed in solution and closely resembles the conformation of a Watson–Crick pair. The polymerase could accommodate a wobble pairing in this position; a previously published G·T mismatch structure (16) adopts a wobble conformation in the postinsertion site that resembles the G·T (and C·O6MeG) solution conformation, with the primer strand base shifted into the major groove. In the polymerase G·T mispair complex, wobble binding causes a concomitant disruption of the template strand, but the template strand is not disrupted in the C·O6MeG pair structure. The T·O6MeG pair in the −1 position resembles the insertion site conformation, with well defined density for the O-methyl group, which again points toward the thymine carbonyl. The position of the O-methyl group, however, is not well defined in the C·O6MeG pair, consistent with it adopting both possible conformations.
Duplex-Binding Region (−2 Base Pair Position).
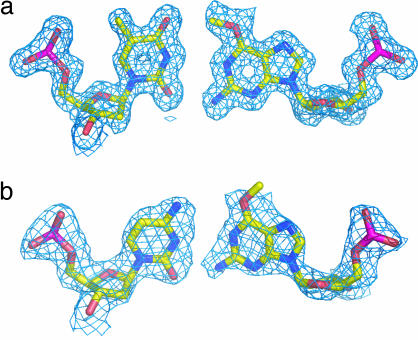
Enzymatic incorporation of a second nucleotide after accurate or mutagenic replication of O6MeG in BF/DNA crystals allows the extension of the DNA to the –2 position (Fig. 2d). In this position, the T·O6MeG pair remains in an isosteric conformation and the methyl conformation is unchanged from that in the active site or the –1 position (Fig. 3a). The C·O6MeG structure is perturbed slightly from an ideal isosteric conformation (Fig. 3b). This may represent a mixture of populations or a single, intermediate conformation; these cannot be distinguished by crystallography at this resolution. In both the C·O6MeG and T·O6MeG −2 complexes, the active site is unperturbed. As in the insertion- and postinsertion-site complexes, the O-methyl group clearly adopts a single conformation pointed toward the thymine carbonyl in a T·O6MeG pair; however, the orientation of this group is not well defined in a C·O6MeG pair.
Fig. 3.
T·O6MeG (a) and C·O6MeG (b) conformations in the −2 base pair position. Mesh shows 1 σ 2 Fo − Fc electron density for the base pairs.
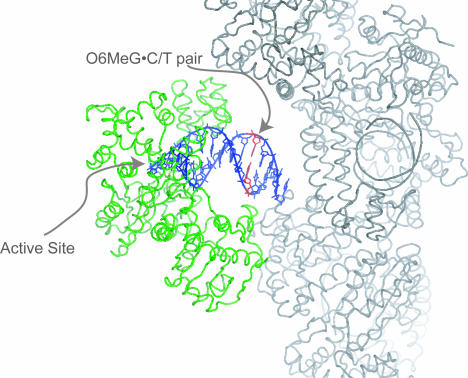
Conformation of T·O6MeG and C·O6MeG Base Pairs in Duplex DNA (−10 Position).
We examined the conformations of O6MeG base pairs unperturbed by protein–DNA interactions with DNA substrates in which either a T·O6MeG or C·O6MeG base pair was located in a large solvent channel of the crystal at the −10 base pair position (Fig. 4). These experiments also allowed us to confirm that the base-pair conformations previously observed at low salt and near-neutral pH match the conformations seen under our crystallization conditions (60% saturated ammonium sulfate and pH 5.8). SI Fig. 7 shows 2 Fo − Fc density for C·O6MeG and T·O6MeG base pairs in the −10 position. The C·O6MeG pair is in a wobble conformation and T·O6MeG pair is in a Watson–Crick isosteric conformation. The conformations of the O6-methyl groups are not well determined in the electron density. Comparison of the crystallographic B factors of DNA with O6MeG·T or O6MeG·C base pairs to B factors from BF/DNA structures with only canonical base pairs shows an increase in disorder in the vicinity of O6MeG. In the O6MeG −10 base pair structures, the disorder propagates to the end of the helix, consistent with the O6MeG causing helix fraying.
Fig. 4.
Observation of O6MeG·T and O6MeG·C in duplex DNA in the solvent channels of BF crystals. The −10 base pair position (red) lies in a solvent channel between BF (green) and its symmetry mates (gray).
Discussion
Both O6MeG Mispairs and Correct Pairs Are Isosteric to Watson–Crick Pairs When Bound by Polymerase.
Polymerases achieve substrate specificity by selecting for dNTPs with complementary shape and hydrogen-bonding patterns to template nucleotides (24, 25, 33–35). One might expect that misinsertion or replication of damaged DNA would be governed by fundamentally similar mechanisms, with substrate preference determined by the extent to which noncanonical interactions mimic the essential steric features of molecular recognition between the polymerase and its preferred substrates. Previously published work on 8-oxo-G miscoding gives an elegant example of such mimicry (21, 36). T·O6MeG and C·O6MeG base pairs each adopt different conformations in their respective polymerase complexes and free-duplex DNA structures. In T·O6MeG complexes, the orientation of the OMe moiety is altered relative to the solution structure. The change from B form to A form DNA upon polymerase binding creates space for the OMe to form a weak but favorable interaction with the thymine carbonyl oxygen. C·O6MeG pairs bound in the polymerase −1 or −2 position adopt a conformation consistent with a tautomer of C that had been observed in C·O6MeG pairs at low pH (Fig. 1). The effect of these changes in conformation or ionization is for the O6-methylated base pairs to assume a shape that approximates that of a canonical Watson–Crick base pair, thereby evading polymerase proofreading mechanisms. O6MeG is a relatively nonspecific base-pairing partner; the ΔΔG of duplex formation between the best and worst O6MeG pairings is <1 kcal/mol, as compared with an ≈5 kcal/mol range for all possible G pairings (28). Therefore, the energetic cost of distortions to O6MeG base pairs presumably is modest enough to be offset by the formation of favorable contacts between the protein and the DNA backbone.
The O6Me Group Forms a Weak Electrostatic Interaction with a Thymine Carbonyl.
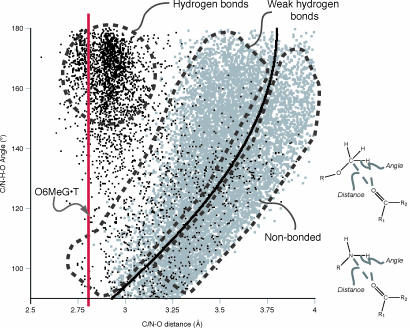
Although O-methyl protons are poor hydrogen bond donors, the O6Me of O6MeG points toward the carbonyl oxygen (O4) of its thymine base-pairing partner in the polymerase active site and in the −1 and −2 positions. A small-molecule crystal structure of O6MeGTP reveals a similar interaction between the OMe protons and the oxygen atom of a solvent water molecule (37). Such weak hydrogen bonds are primarily electrostatic in character and generally provide <1 kcal/mol of stabilization (38). A comparison between O-methyl:carbonyl and amine:carbonyl interactions from the Cambridge Structural Database (39) shows that both interactions are common but they differ in their geometry (Fig. 5). Although we cannot directly observe protons in our crystal structure, the observed methyl carbon-carbonyl distance is consistent with a 2.75-Å H-O distance based on the trends seen in the Cambridge Structural Database (Fig. 5). This methyl-proton:carbonyl interaction was not observed in previous crystallographic studies of B form DNA duplexes containing T·O6MeG pairs in the absence of protein (30). The BF polymerase binds DNA in an A form conformation with significant base pair opening (SI Table 4), providing additional space to accommodate the methyl group.
Fig. 5.
Geometry of carbonyl oxygen:O-methyl interactions from small-molecule crystal structures in the Cambridge Structural Database (39). Gray dots, OMe proton–carbonyl interactions with H-O distances <4.5 Å, and O-H-O angles of 90° or greater were selected from the Cambridge Structural Database, yielding 9,418 interacting pairs. Black dots, amine–carbonyl interactions were selected in the same manner, yielding 3,151 interacting pairs. O-methyl-carbon or amine nitrogen to carbonyl oxygen interatomic distances are plotted vs. the O/N-H-carbonyl oxygen angle; these parameters are depicted schematically beside the graph. Interactions above and to the left of the heavy line have proton-carbonyl interatomic distances less than the sum of the oxygen and hydrogen van der Waals radii, i.e., they can be defined as bonded. The plot is marked to indicate general locations of hydrogen bonded, weak hydrogen bonded, and nonbonded interactions. The average interaction distance for O6MeG·T pairs in the insertion site, −1, and −2 position structures is indicated with a red vertical line at 2.80 Å.
O6MeG Is a Promiscuous Base-Pairing Partner, Which Allows it to Incorporate T and C Indiscriminately.
High-fidelity polymerases incorporate T and C opposite O6MeG to a similar extent despite structural differences in the solution conformations of O6MeG·T and O6MeG·C base pairs. This study reveals that O6MeG is a relatively plastic base-pairing partner and that polymerase binding induces changes in both C·O6MeG and T·O6MeG base pairs. The energetic barrier to forming a C·O6MeG isosteric pair is overcome by the DNA polymerase but likely accounts for the preferential insertion of T opposite O6MeG by high-fidelity polymerases. On the other hand, a lower fidelity polymerase like pol η with a relaxed shape requirement does not require that C·O6MeG form an isosteric pair and, thus, can insert C more frequently (12).
The notion that mutagenic DNA replication occurs because transient base pair conformational switching of mispaired nucleotides allows them to mimic canonical base pairs has been put forth since 1953 (40, 41). Replication fidelity, however, results from a complex interaction between the DNA polymerase and its substrates. In the absence of detailed structural characterization of relevant polymerase–DNA complexes, the precise mechanism by which mimicry of canonical base pairs is achieved, or even whether such mimicry occurs, has been the subject of much debate. Our observations of O6MeG mispairs in Watson–Crick-like conformations in the polymerase active site and the −1 and −2 base pair positions provide direct experimental evidence to support the hypothesis that mutagenic replication can occur when polymerase binding alters base-pair conformations to resemble those of Watson–Crick pairs.
Methods
Protein Preparation.
Wild-type and mutant proteins were purified as described in ref. 42. Wild-type protein was used for all kinetics experiments and all crystallizations except ddTTP/ddCTP:BF:DNA ternary complexes, which were grown by using BF D598/F710Y. Use of the D598A mutation to destabilize a crystal contact that otherwise traps BF–DNA complex crystals in an open form has been described in ref. 17. The F710Y mutation was designed based on homologous E. coli and phage T7 DNA polymerase I mutations, which decrease discrimination against dideoxynucleotides (43, 44). The QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) was used to construct the double mutant F710Y/D598A from the D598A mutant gene.
Cocrystallization of BF with DNA Primer Template and dNTP Substrates.
Complementary oligonucleotides (Midland Certified Reagent Co., Midland, TX) (+1 complex, 5′-CAT(O6MeG)CGAGTCAGG-3′ and 5′-CTGACTCG-3′; −1 and −2 complexes, 5′-GTAC(O6MeG)AGCTGATCGCA-3′ and 5′-GCGATCAGC-3′; closed ternary complexes, 5′-CAT(O6MeG)CGAGTCAGG-3′ and 5′-CCTGACTC-3′; −10 position, 5′-CGGCCTGACTCG(C/T)ATGA and 5′-CAT(O6MeG)CGAGTCAGG-3′) were annealed as described in ref. 17. The resulting DNA substrates contain an 8- to 13-bp duplex region, a 5′ single-stranded template overhang, and, in the case of the +1, −1, −2, and −10 substrates, a single-nucleotide 3′ template overhang. The 3′ overhang prevents DNA from binding backward (17). Ternary complexes, however, crystallize more readily with a blunt end; two noncrystallographic-symmetry-related DNA molecules stack pseudocontinuously in the crystals. Use of this substrate also eliminates merohedral twinning seen in previous closed complex structures (17). An analogous ternary complex crystal with a ddCTP·G pair was solved to allow direct comparison to these O6MeG ternary complex structures, see SI Methods and SI Table 3. Binary BF–DNA complex crystals were obtained as described in ref. 17. −1 and −2 position complexes were generated from +1 complex crystals by catalysis in the crystal (see below). A C·O6MeG −1 binary complex solved at pH 7.3 is indistinguishable from the complex presented in this study. BF:DNA:ddTTP ternary complex was obtained by incubating the D598A/F710Y double mutant with appropriate DNA at a 1:3 molar ratio in 3.5 mM ddGTP, 3.5 mM ddTTP, 10 mM MgSO4, and 10 mM MnSO4. A BF DNA ddCTP:O6MeG ternary complex was obtained by soaking dTTP complex crystals overnight in 60% saturated ammonium sulfate/2.5% 2-methyl-2,4-pentanediol/100 mM Mes, pH 5.8, which serves to eliminate all bound ddTTP and then soaking overnight in the same solution supplemented with 100 mM MnSO4/100 mM ddCTP. Ternary complex crystals were grown as described for binary complex crystals in ref. 17, with the addition of 10 mM MnSO4.
Data Collection, Structure Determination, and Analysis.
Diffraction data were collected by using a Rigaku rotating anode x-ray generator or at the Advanced Photon Source (see SI Table 3). Data were processed by using HKL2000 (45) or XDS (46). Rigid body refinement of previously published complexes provided the starting phases for refinement. The ternary complex was solved by molecular replacement in CNS (47) with our previously published closed complex (17) as a search model. Crystallographic refinement was carried out with CNS (47) or REFMAC5 (48), and model rebuilding was carried out in O (49) or Coot (50). Final models had reasonable geometry, with bond angle and length rmsd values of 1.23 ± 0.09° and 0.006 ± 0.002 Å, respectively (see Table 1 and SI Table 3). Suitable topology and parameter files for O6MeG, ddTTP, and ddCTP were generated with PRODRG (51); however, the C6-O6-Me angle was set to 116.5°, which better agrees with the published O6MeG small molecule structure (37) and our electron density. Polymerase structures were superimposed by using the Cα atoms of the palm subdomain (residues 646–655, 823–838, and 863–869).
Catalysis in the Crystal.
Polymerase activity was initiated by incubating crystals in stabilization solutions containing 30 mM total dNTP(s) and 60 mM MgSO4 for 24 h at 17°C.
Determination of BF Kinetic Parameters.
Kinetic parameters were determined as described in ref. 32. Briefly, radiolabeled primer (5′-GTGCCTGACTCG) was annealed to a DNA template [5′-CAT(G/O6MeG)CGAGTCAGGCACT]. DNA–protein complexes were preformed in reaction buffer (50 mM Tris·HCl, pH 8.0/5 mM MgSO4/2.5% glycerol/10 nM DTT/100 μg/ml BSA). Reactions were initiated by mixing with varying concentrations of dCTP or dTTP (also in reaction buffer) for various times and then quenched by addition of EDTA to a final concentration of 300 mM. Time points of 2 seconds or longer were taken on the benchtop. Shorter timepoints were produced by using a pulsed-quench-flow apparatus (KinTek, State College, PA). All experiments were conducted at room temperature. Products were separated by gel electrophoresis and quantitated by a phosphorimager.
Supplementary Material
Acknowledgments
We thank H. W. Hellinga for critical reading of the manuscript and T. Pohlhaus and A. Changela for assistance with data collection. Data for this study were measured at Southeast Regional Collaborative Access Team (SER-CAT), and Structural Biology Center beamlines were measured at the Advanced Photon Source, Argonne National Laboratory. SER-CAT supporting institutions may be found at www.ser-cat.org/members.html. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This work was supported by the Structural Cell Biology of DNA Repair Program Grant 5 P01 CA92584 from the National Cancer Institute (to L.S.B.) and an Agouron Institute Fellowship from the Jane Coffin Childs Memorial Fund for Cancer Research (to J.J.W.).
Abbreviations
- BF
Bacillus stearothermophilus DNA polymerase I, large fragment
- O6MeG
O6-methyl-guanine.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2HHQ, 2HHS, 2HHT, 2HHU, 2HHV, 2HHW, 2HW3, 2HHX, 2HVH, and 2HVI).
This article contains supporting information online at www.pnas.org/cgi/content/full/0609580103/DC1.
References
- 1.Dipple A. Carcinogenesis. 1995;16:437–441. doi: 10.1093/carcin/16.3.437. [DOI] [PubMed] [Google Scholar]
- 2.Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. DNA Repair. 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 4.Rydberg B, Lindahl T. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taverna P, Sedgwick B. J Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loveless A. Nature. 1969;223:206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- 7.Newbold RF, Warren W, Medcalf AS, Amos J. Nature. 1980;283:596–599. doi: 10.1038/283596a0. [DOI] [PubMed] [Google Scholar]
- 8.Dumenco LL, Allay E, Norton K, Gerson SL. Science. 1993;259:219–222. doi: 10.1126/science.8421782. [DOI] [PubMed] [Google Scholar]
- 9.Loechler EL, Green CL, Essigmann JM. Proc Natl Acad Sci USA. 1984;81:6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green CL, Loechler EL, Fowler KW, Essigmann JM. Proc Natl Acad Sci USA. 1984;81:13–17. doi: 10.1073/pnas.81.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodside AM, Guengerich FP. Biochemistry. 2002;41:1027–1038. doi: 10.1021/bi011495n. [DOI] [PubMed] [Google Scholar]
- 12.Haracska L, Prakash S, Prakash L. Mol Cell Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singer B, Chavez F, Goodman MF, Essigmann JM, Dosanjh MK. Proc Natl Acad Sci USA. 1989;86:8271–8274. doi: 10.1073/pnas.86.21.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dosanjh MK, Galeros G, Goodman MF, Singer B. Biochemistry. 1991;30:11595–11599. doi: 10.1021/bi00113a015. [DOI] [PubMed] [Google Scholar]
- 15.Woodside AM, Guengerich FP. Biochemistry. 2002;41:1039–1050. doi: 10.1021/bi011496f. [DOI] [PubMed] [Google Scholar]
- 16.Johnson SJ, Beese LS. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 17.Johnson SJ, Taylor JS, Beese LS. Proc Natl Acad Sci USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiefer JR, Mao C, Braman JC, Beese LS. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 19.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. J Biol Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 20.Hsu GW, Kiefer JR, Burnouf D, Becherel OJ, Fuchs RP, Beese LS. J Biol Chem. 2004;279:50280–50285. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 21.Hsu GW, Ober M, Carell T, Beese LS. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 22.Johnson KA. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 23.Kunkel TA, Bebenek K. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 24.Kool ET. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 25.Goodman MF, Creighton S, Bloom LB, Petruska J. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 26.Joyce CM, Benkovic SJ. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 27.Patel DJ, Shapiro L, Kozlowski SA, Gaffney BL, Jones RA. Biochemistry. 1986;25:1027–1036. doi: 10.1021/bi00353a012. [DOI] [PubMed] [Google Scholar]
- 28.Gaffney BL, Jones RA. Biochemistry. 1989;28:5881–5889. doi: 10.1021/bi00440a026. [DOI] [PubMed] [Google Scholar]
- 29.Patel DJ, Shapiro L, Kozlowski SA, Gaffney BL, Jones RA. Biochemistry. 1986;25:1036–1042. doi: 10.1021/bi00353a013. [DOI] [PubMed] [Google Scholar]
- 30.Leonard GA, Thomson J, Watson WP, Brown T. Proc Natl Acad Sci USA. 1990;87:9573–9576. doi: 10.1073/pnas.87.24.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan HB, Swann PF, Chance EM. Biochemistry. 1994;33:5335–5346. doi: 10.1021/bi00183a042. [DOI] [PubMed] [Google Scholar]
- 32.Johnson KA. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- 33.Carroll SS, Cowart M, Benkovic SJ. Biochemistry. 1991;30:804–813. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- 34.Echols H, Goodman MF. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 35.Kunkel TA. J Biol Chem. 1992;267:18251–18254. [PubMed] [Google Scholar]
- 36.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parthasarathy R, Fridey SM. Carcinogenesis. 1986;7:221–227. doi: 10.1093/carcin/7.2.221. [DOI] [PubMed] [Google Scholar]
- 38.Desiraju GR, Steiner T. The Weak Hydrogen Bond in Structural Chemistry and Biology. Oxford: Oxford Univ Press/Int Union Crystallogr; 1999. [Google Scholar]
- 39.Allen FH. Acta Crystallogr B. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 40.Watson JD, Crick FHC. Cold Spring Harbor Symp Quant Biol. 1953;18:123–129. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- 41.Topal MD, Fresco JR. Nature. 1976;263:285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- 42.Kiefer JR, Mao C, Hansen CJ, Basehore SL, Hogrefe HH, Braman JC, Beese LS. Structure (London) 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 43.Astatke M, Grindley NDF, Joyce CM. J Mol Biol. 1998;278:147–165. doi: 10.1006/jmbi.1998.1672. [DOI] [PubMed] [Google Scholar]
- 44.Tabor S, Richardson CC. Proc Natl Acad Sci USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Kabsch W. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 47.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.vanAalten DMF, Bywater R, Findlay JBC, Hendlich M, Hooft RWW, Vriend G. J Comput Aided Mol Des. 1996;10:255–262. doi: 10.1007/BF00355047. [DOI] [PubMed] [Google Scholar]
- 52.Gaffney BL, Goswami B, Jones RA. J Am Chem Soc. 1993;115:12607–12608. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.