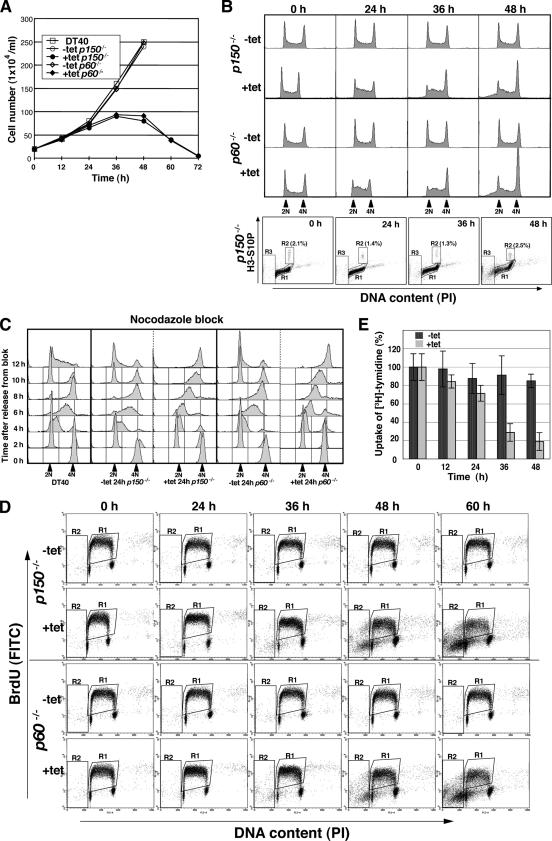
Figure 2.
Growth curves and cell cycle analyses of p150- and p60-conditional knockout cells. (A) Growth curves for DT40, and p150- and p60-conditional knockout cells. The numbers of indicated genotype cells in the absence (−tet) or presence (+tet) of tet were counted. Each time point was examined in triplicate. (B) Flow cytometric analysis of asynchronous p150- and p60-conditional knockout cells. Asynchronous p150 (two top panels) and p60 (two middle panels) conditional cells were fixed and stained with PI to detect total DNA (x-axis; linear scale) at indicated time points in the absence (−tet) and presence (+tet) of tet. Bottom, cells were fixed, stained with anti-histone H3 phosphorylated at Ser10 antibody (H3-S10P) followed by a secondary AlexaFluor 488-labeled anti-rabbit antibody (y-axis; log scale), and finally counterstained with PI to detect total DNA (x-axis; linear scale). Boxed regions R1, R2, and R3 represent interphase, mitotic and apoptotic cells, respectively. (C) Flow cytometric analysis of synchronized p150- and p60-conditional knockout cells. For synchronization into mitotic phase, cells were cultured in the presence (+tet 24 h) or absence of tet (−tet 24 h) for 17 h, followed by culture with nocodazole (500 ng/ml) for 7 h. After release from the cell cycle block, cells were collected at 2 h intervals, fixed, and stained with PI to detect total DNA (x-axis; linear scale). (D) Two-dimensional flow cytometric analysis of p150- and p60-conditional knockout cells. At indicated times, p150 (two top panels) and p60 (two bottom panels) conditional cells were pulse labeled with BrdU for 10 min, fixed, stained with FITC-labeled anti-BrdU antibody (y-axis; log scale), and then stained with PI to detect total DNA (x-axis, linear scale). Boxed regions R1 and R2 indicate BrdU-incorporated S phase and apoptotic cells, respectively. (E) Relative DNA synthesis rate following p150 depletion. The [3H]thymidine incorporations at indicated times after cultivation with (+tet) or without (−tet) tet were normalized by dividing total cell numbers at each time point by those at 0 h, defined as 100%. Data are from two separate experiments.