Abstract
Three-dimensional (3D) structure is now known for a large fraction of all protein families. Thus, it has become rather likely that one will find a homolog with known 3D structure when searching a sequence database with an arbitrary query sequence. Depending on the extent of similarity, such neighbor relationships may allow one to infer biological function and to identify functional sites such as binding motifs or catalytic centers. Entrez's 3D-structure database, the Molecular Modeling Database (MMDB), provides easy access to the richness of 3D structure data and its large potential for functional annotation. Entrez's search engine offers several tools to assist biologist users: (i) links between databases, such as between protein sequences and structures, (ii) pre-computed sequence and structure neighbors, (iii) visualization of structure and sequence/structure alignment. Here, we describe an annotation service that combines some of these tools automatically, Entrez's ‘Related Structure’ links. For all proteins in Entrez, similar sequences with known 3D structure are detected by BLAST and alignments are recorded. The ‘Related Structure’ service summarizes this information and presents 3D views mapping sequence residues onto all 3D structures available in MMDB (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=structure).
CONTENT
Access
The molecular modeling database (MMDB) is Entrez's ‘Structure’ database (1). Querying MMDB with text terms, e.g. one may identify structures of interest based on a protein name. Links between databases provide other search mechanisms. A query of Entrez PubMed database, e.g. will identify articles citing a particular protein name. Links from this set of articles to ‘Structure’ may identify structures not found by direct query, since PubMed abstracts contain additional descriptive terms. Currently, MMDB and its visualization services handle ∼25 000 user queries per day.
Data sources
Experimental three-dimensional (3D) structure data are obtained from the Protein Data Bank (PDB) (2). Author-annotated features provided by PDB are recorded in MMDB. The agreement between atomic coordinate and sequence data is verified, and sequence data are obtained from PDB coordinate records, if necessary, to resolve ambiguities(3). Data are mapped into a computer friendly format and transferred between applications using Abstract Syntax Notation 1 (ASN.1). This validation and encoding supports the interoperable display of sequence, structure and alignment. Uniformly defined secondary-structure and 3D-domain features are added to support structure neighbor calculations. MMDB currently contains ∼39 000 structure entries, corresponding to ∼90 000 chains and 170 000 3D domains.
Summary, links, neighbors and visualization
The MMDB web server generates structure summary pages, which provide a concise description of an MMDB entry's content and the available annotation (4). Sequences derived from MMDB are entered into Entrez's protein or nucleic acid sequence database, preserving links to the corresponding 3D structures. Links to PubMed are generated by matching citations. Links to Entrez's organism taxonomy database are generated by semi-automatic processing of ‘source records' and other descriptive text provided by PDB. Ligands and other small molecules are identified and added to the PubChem resource, accessible at http://pubchem.ncbi.nlm.nih.gov, also preserving reciprocal links to 3D structure. Sequence neighbors are identified by BLAST (5), and links to the Conserved Domain Database (CDD) (6) by the RPS-BLAST algorithm (5). Structure neighbors are identified by VAST (7). The 3D structure viewer supported by Entrez, Cn3D (8), provides molecular-graphics visualization.
ANNOTATING SEQUENCE WITH STRUCTURE
The ‘Related Structure’ service
In the Entrez database system, protein sequences are neighbored to each other by comparing each newly entered sequence to all other database entries. These database scans are run with the BLAST (5) engine, which identifies sequence neighbors with significant similarity, and the resulting sequence identifiers and taxonomy indices are stored, so that Entrez can provide ‘Related Sequences’ links for all protein records in the collection. The ‘Related Structure’ service is built on top of this system. Sequence neighbors directly linked to MMDB are identified and alignments are re-computed by employing the ‘BlastTwoSequences’ tool (9) to restore alignment footprints. The ‘Related Structure’ web interface provides direct access to this information. Initially this service had been restricted to sequences from microbial genomes (10), but it has now been expanded to cover all proteins in Entrez and is updated daily to provide a comprehensive 3D-structure annotation service. Identification of structure-linked neighbors and the visualization of sequence-structure alignment is also possible using Entrez and the Cn3D alignment viewer/editor, but ‘Related Structures’ provides a convenient new summary and ‘one click’ shortcuts to 3D visualization. These 3D views may be used to identify conserved residues and map site-specific features derived from the 3D structure. Currently ∼48% of non-identical protein sequences in Entrez have been linked to at least one related structure, employing a conservative threshold for alignment length (50 aligned residues or more) and similarity (30% or more identical residues in the aligned footprint); see Figure 1 for details.
Figure 1.
Non-identical protein sequences in Entrez have been classified into groups linked to related structures, at various levels of sequence similarity. Sequence identity is calculated from the BLAST alignments, and here only those neighbor relationships are listed that produce an aligned footprint of 50 residues or more. The analysis also excludes protein sequences which have been directly obtained from MMDB. Forty-eight percent of sequences in Entrez protein have at least one structure neighbor with an extensive alignment footprint and at least 30% identical residues.
An example
A search with the term ‘Angiotensin converting enzyme’ in Entrez's protein database retrieves >400 hits. One may configure the Entrez browser to filter search results by various criteria, and one pre-configured filter selects those protein sequences with ‘Related Structures’ (configuration of Entrez can be achieved by following links to ‘My NCBI’, or by clicking on the ‘toolbox’ icon shown at the top of Entrez document summaries.). In this example, the ‘Related Structures’ filter shows that >240 of the identified sequence records have links to related structures.
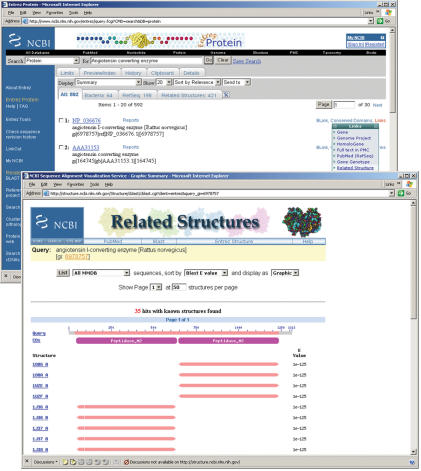
One such protein sequence is the ACE protein from Rattus norvegicus (accession no. ‘NP_036676’). On the ‘Links’ menu for this record, ‘Related structures’ generates a request to the Related Structure service (http://structure.ncbi.nlm.nih.gov/Structure/cblast/cblast.cgi?client=entrez&query_gi=6978757). The resulting page indicates with a horizontal bar, the sequence region annotated by each related structure (Figure 2). The display also supports sorting by a variety of alignment parameters such as score or length and selection of sequence-dissimilar ‘non redundant’ subsets. A ‘Table’ option switches to a text view, listing descriptions of each structure as well as alignment scores.
Figure 2.
A screen shot of the ‘Related Structure’ summary along with Entrez's document summary for protein NP_036676. Clicking on the ‘Related Structure’ option from the ‘Links’ pull-down menu launches the summary view.
Using the table view with this example, one may notice that several related structures are complexes of the same protein with different drugs/inhibitors, e.g. structures with PDB codes 1O86 (11), 1UZF (12) and 1UZE (12). Clicking on the graphical alignment footprint of 1O86, a human ACE enzyme in complex with lisinopril, one can see a text representation of the corresponding BLAST alignment, and a Cn3D view of the alignment can be launched by clicking on ‘Get 3D Structure data’ (Figure 3). One may see that the query protein is highly similar in sequence to the human ACE enzyme, as identical residue pairs are colored red by default. The sequence identity across the aligned region is 82%, and it appears that the core of the structure is mostly formed by residues conserved between the two aligned rows, while non-conserved residues are mainly located on the structure's surface.
Figure 3.
A Cn3D view of the query sequence from Figure 2 aligned to chain A of the related structure 1O86 (PDB code). Residues in aligned regions are displayed in upper case letters with identical residue pairs rendered in red color. Residues within a 5 A contact radius of the bound drug lisinopril are highlighted in the 3D structure view and automatically mapped onto the aligned residues shown in the sequence alignment window. Side chains of these residues are displayed selectively and rendered as ball-and-stick models.
One may further identify the catalytic center by identifying residues that contact the catalytic Zinc ion. Those sites can then be mapped from the structure to aligned regions in the sequence window using Cn3D's highlighting functionality. One may also examine the sequence-structure alignments with related structures 1UZE and 1UZF, human ACE binding to enalaprilat and captopril, respectively, drugs with chemical structures similar to that of lisinopril. This allows one to identify conserved interactions between the ACE enzyme and this series of antihypertensive drugs. Similarly, by examining the related structure 2AJF (13), one may be able to identify residues critical for cross-species infection by studying the protein–protein interactions between the receptor binding domain from SARS Coronavirus Spike and human versus rat angiotensin-converting enzyme 2.
The ‘Related Structure’ service is also integrated with NCBI's protein BLAST service. A ‘Related Structures’ link is provided when one or more similar proteins with known 3D structures have been identified by BLAST. The NCBI single-nucleotide polymorphism resource (SNP) also links to the ‘Related Structure’ service, which in this context provides a mapping of both synonymous and non-synonymous coding SNPs onto experimentally determined 3D structures. ‘Related Structure’ may be expanded further in the future, to provide visualization for other NCBI resources and to support additional filtering and selection among related structures, e.g. to highlight those annotated with conserved domain footprints by the CDD resource or those linked to small molecules in the PubChem database.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by US government.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wheeler D.L., Barrett T., Benson D.A., Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deshpande N., Addess K.J., Bluhm W.F., Merino-Ott J.C., Townsend-Merino W., Zhang Q., Knezevich C., Xie L., Chen L., Feng Z., et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohkawa H., Ostell J., Bryant S. MMDB: an ASN.1 specification for macromolecular structure. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:259–667. [PubMed] [Google Scholar]
- 4.Chen J., Anderson J.B., DeWeese-Scott C., Fedorova N.D., Geer L.Y., He S., Hurwitz D.I., Jackson J.D., Jacobs A.R., Lanczycki C.J., et al. MMDB: Entrez's 3D-structure database. Nucleic Acids Res. 2003;31:474–477. doi: 10.1093/nar/gkg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchler-Bauer A., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibrat J.F., Madej T., Bryant S.H. Surprising similarities in structure comparison. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Geer L.Y., Chappey C., Kans J.A., Bryant S.H. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 9.Tatusova T.A., Madden T.L. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Bryant S., Tatusov R., Tatusova T. Links from genome proteins to known 3D structures. Genome Res. 2000;10:1643–1647. doi: 10.1101/gr.143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Natesh R., Schwager S.L., Sturrock E.D., Acharya K.R. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature. 2003;421:551–554. doi: 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- 12.Natesh R., Schwager S.L., Evans H.R., Sturrock E.D., Acharya K.R. Structural details on the binding of antihypertensive drugs captopril and enalaprilat to human testicular angiotensin I-converting enzyme. Biochemistry. 2004;43:8718–8724. doi: 10.1021/bi049480n. [DOI] [PubMed] [Google Scholar]
- 13.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1822–1823. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]